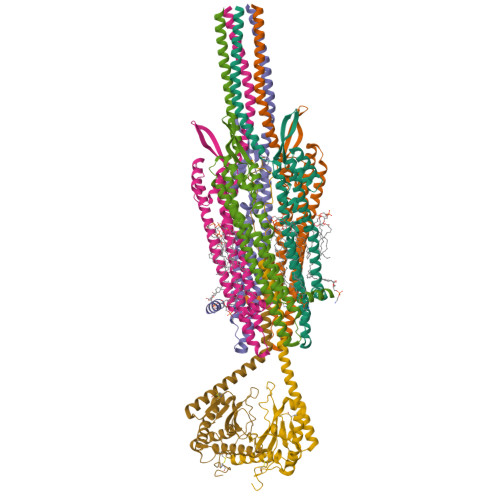
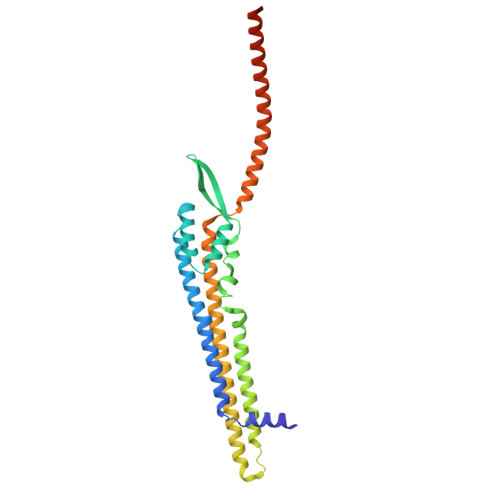
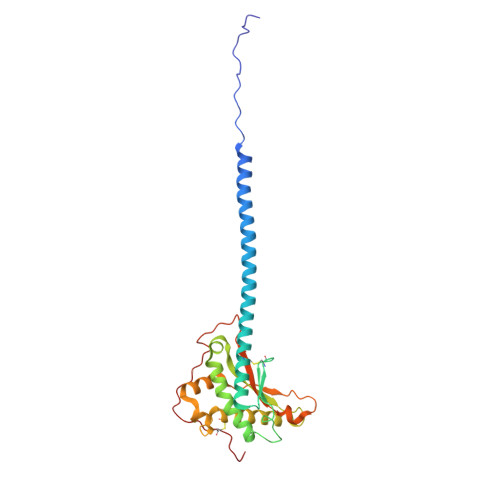
Structure and mechanism of the Zorya anti-phage defence system.
Hu, H., Popp, P.F., Hughes, T.C.D., Roa-Eguiara, A., Rutbeek, N.R., Martin, F.J.O., Hendriks, I.A., Payne, L.J., Yan, Y., Humolli, D., Klein-Sousa, V., Songailiene, I., Wang, Y., Nielsen, M.L., Berry, R.M., Harms, A., Erhardt, M., Jackson, S.A., Taylor, N.M.I.(2025) Nature 639: 1093-1101
- PubMed: 39662505
- DOI: https://doi.org/10.1038/s41586-024-08493-8
- Primary Citation of Related Structures:
8QY7, 8QYC, 8QYD, 8QYH, 8QYK, 8QYY, 8R68 - PubMed Abstract:
Zorya is a recently identified and widely distributed bacterial immune system that protects bacteria from viral (phage) infections. Three Zorya subtypes have been discovered, each containing predicted membrane-embedded ZorAB complexes paired with soluble subunits that differ among Zorya subtypes, notably ZorC and ZorD in type I Zorya systems 1,2 . Here, we investigate the molecular basis of Zorya defense using cryo-electron microscopy, mutagenesis, fluorescence microscopy, proteomics, and functional studies. We present cryo-EM structures of ZorAB and show that it shares stoichiometry and features of other 5:2 inner membrane ion-driven rotary motors. The ZorA 5 B 2 complex contains a dimeric ZorB peptidoglycan binding domain and a pentameric α-helical coiled-coil tail made of ZorA that projects approximately 70 nm into the cytoplasm. We also characterize the structure and function of the soluble Zorya components, ZorC and ZorD, finding that they harbour DNA binding and nuclease activity, respectively. Comprehensive functional and mutational analyses demonstrate that all Zorya components work in concert to protect bacterial cells against invading phages. We provide evidence that ZorAB operates as a proton-driven motor that becomes activated upon sensing of phage invasion. Subsequently, ZorAB transfers the phage invasion signal through the ZorA cytoplasmic tail to recruit and activate the soluble ZorC and ZorD effectors, which facilitate degradation of the phage DNA. In summary, our study elucidates the foundational mechanisms of Zorya function as an anti-phage defense system.
Organizational Affiliation:
Structural Biology of Molecular Machines Group, Protein Structure & Function Program, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark. haidai.hu@cpr.ku.dk.