A macrocyclic kinase inhibitor overcomes triple resistant mutations in EGFR-positive lung cancer.
Suzuki, M., Uchibori, K., Oh-Hara, T., Nomura, Y., Suzuki, R., Takemoto, A., Araki, M., Matsumoto, S., Sagae, Y., Kukimoto-Niino, M., Kawase, Y., Shirouzu, M., Okuno, Y., Nishio, M., Fujita, N., Katayama, R.(2024) NPJ Precis Oncol 8: 46-46
- PubMed: 38396251
- DOI: https://doi.org/10.1038/s41698-024-00542-9
- Primary Citation of Related Structures:
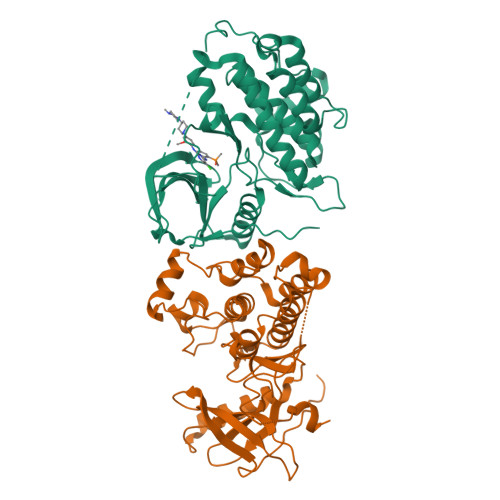
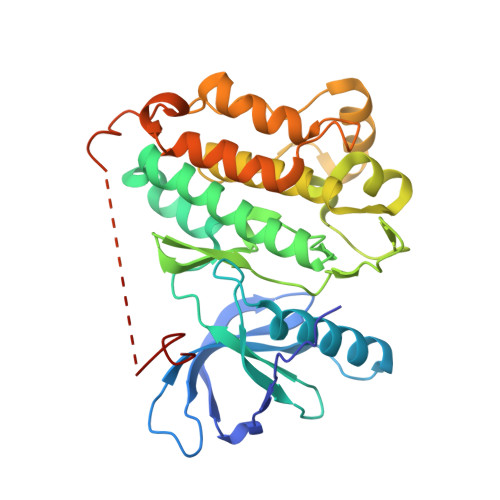
8H7X - PubMed Abstract:
Brigatinib-based therapy was effective against osimertinib-resistant EGFR C797S mutants and is undergoing clinical studies. However, tumor relapse suggests additional resistance mutations might emerge. Here, we first demonstrated the binding mode of brigatinib to the EGFR-T790M/C797S mutant by crystal structure analysis and predicted brigatinib-resistant mutations through a cell-based assay including N-ethyl-N-nitrosourea (ENU) mutagenesis. We found that clinically reported L718 and G796 compound mutations appeared, consistent with their proximity to the binding site of brigatinib, and brigatinib-resistant quadruple mutants such as EGFR-activating mutation/T790M/C797S/L718M were resistant to all the clinically available EGFR-TKIs. BI-4020, a fourth-generation EGFR inhibitor with a macrocyclic structure, overcomes the quadruple and major EGFR-activating mutants but not the minor mutants, such as L747P or S768I. Molecular dynamics simulation revealed the binding mode and affinity between BI-4020 and EGFR mutants. This study identified potential therapeutic strategies using the new-generation macrocyclic EGFR inhibitor to overcome the emerging ultimate resistance mutants.
Organizational Affiliation:
Division of Experimental Chemotherapy, Cancer Chemotherapy Center, Japanese Foundation for Cancer Research, Tokyo, 135-8550, Japan.