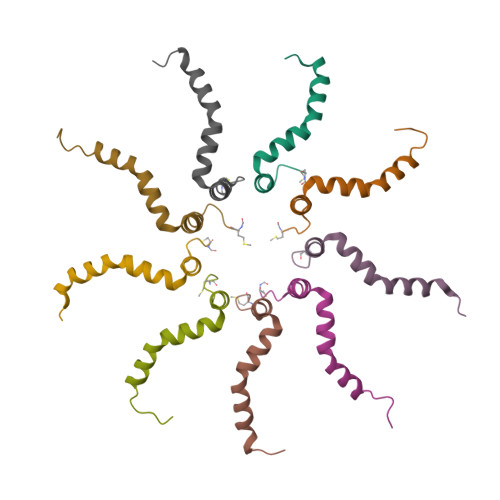
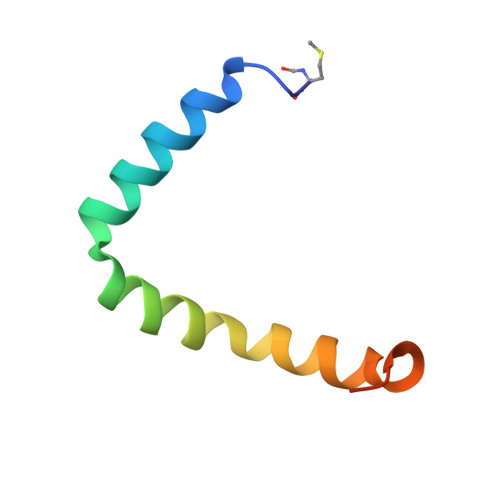
Structural model for the protein-translocating element of the twin-arginine transport system.
Rodriguez, F., Rouse, S.L., Tait, C.E., Harmer, J., De Riso, A., Timmel, C.R., Sansom, M.S., Berks, B.C., Schnell, J.R.(2013) Proc Natl Acad Sci U S A 110: E1092-E1101
- PubMed: 23471988
- DOI: https://doi.org/10.1073/pnas.1219486110
- Primary Citation of Related Structures:
2LZR, 2LZS - PubMed Abstract:
The twin-arginine translocase (Tat) carries out the remarkable process of translocating fully folded proteins across the cytoplasmic membrane of prokaryotes and the thylakoid membrane of plant chloroplasts. Tat is required for bacterial pathogenesis and for photosynthesis in plants. TatA, the protein-translocating element of the Tat system, is a small transmembrane protein that assembles into ring-like oligomers of variable size. We have determined a structural model of the Escherichia coli TatA complex in detergent solution by NMR. TatA assembly is mediated entirely by the transmembrane helix. The amphipathic helix extends outwards from the ring of transmembrane helices, permitting assembly of complexes with variable subunit numbers. Transmembrane residue Gln8 points inward, resulting in a short hydrophobic pore in the center of the complex. Simulations of the TatA complex in lipid bilayers indicate that the short transmembrane domain distorts the membrane. This finding suggests that TatA facilitates protein transport by sensitizing the membrane to transient rupture.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, Oxford, OX1 3QU, United Kingdom.