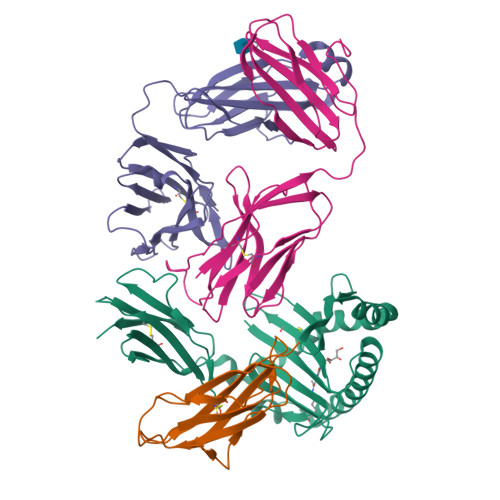
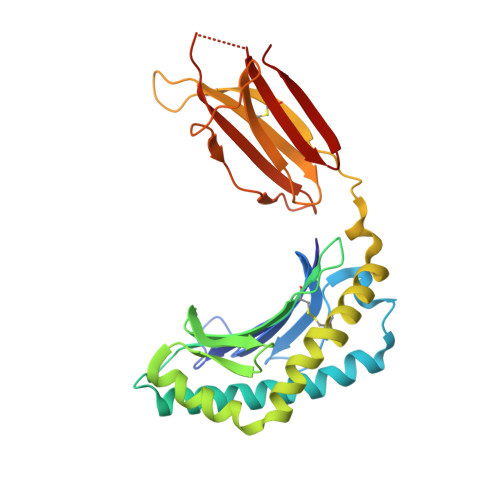
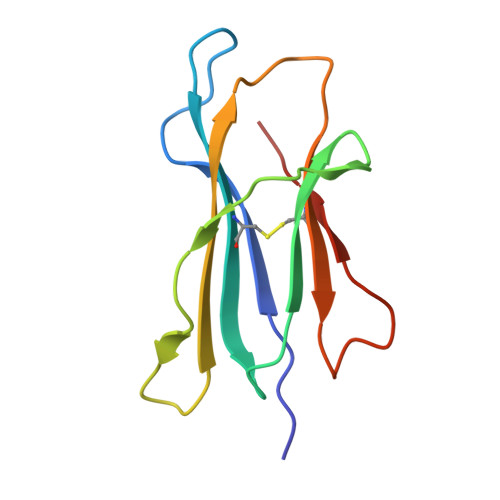
A class of gamma delta T cell receptors recognize the underside of the antigen-presenting molecule MR1.
Le Nours, J., Gherardin, N.A., Ramarathinam, S.H., Awad, W., Wiede, F., Gully, B.S., Khandokar, Y., Praveena, T., Wubben, J.M., Sandow, J.J., Webb, A.I., von Borstel, A., Rice, M.T., Redmond, S.J., Seneviratna, R., Sandoval-Romero, M.L., Li, S., Souter, M.N.T., Eckle, S.B.G., Corbett, A.J., Reid, H.H., Liu, L., Fairlie, D.P., Giles, E.M., Westall, G.P., Tothill, R.W., Davey, M.S., Berry, R., Tiganis, T., McCluskey, J., Pellicci, D.G., Purcell, A.W., Uldrich, A.P., Godfrey, D.I., Rossjohn, J.(2019) Science 366: 1522-1527
- PubMed: 31857486
- DOI: https://doi.org/10.1126/science.aav3900
- Primary Citation of Related Structures:
6MWR - PubMed Abstract:
T cell receptors (TCRs) recognize antigens presented by major histocompatibility complex (MHC) and MHC class I-like molecules. We describe a diverse population of human γδ T cells isolated from peripheral blood and tissues that exhibit autoreactivity to the monomorphic MHC-related protein 1 (MR1). The crystal structure of a γδTCR-MR1-antigen complex starkly contrasts with all other TCR-MHC and TCR-MHC-I-like complex structures. Namely, the γδTCR binds underneath the MR1 antigen-binding cleft, where contacts are dominated by the MR1 α3 domain. A similar pattern of reactivity was observed for diverse MR1-restricted γδTCRs from multiple individuals. Accordingly, we simultaneously report MR1 as a ligand for human γδ T cells and redefine the parameters for TCR recognition.
Organizational Affiliation:
Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria 3800, Australia.