Inside out Porphyridium cruentum : Beyond the Conventional Biorefinery Concept.
Liberti, D., Imbimbo, P., Giustino, E., D'Elia, L., Ferraro, G., Casillo, A., Illiano, A., Pinto, G., Di Meo, M.C., Alvarez-Rivera, G., Corsaro, M.M., Amoresano, A., Zarrelli, A., Ibanez, E., Merlino, A., Monti, D.M.(2023) ACS Sustain Chem Eng 11: 381-389
- PubMed: 36643001
- DOI: https://doi.org/10.1021/acssuschemeng.2c05869
- Primary Citation of Related Structures:
8B4N - PubMed Abstract:
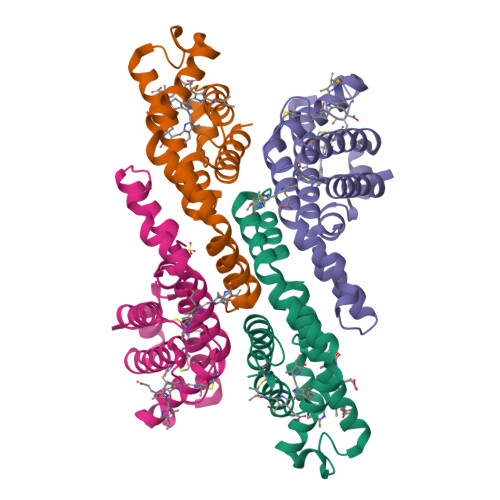
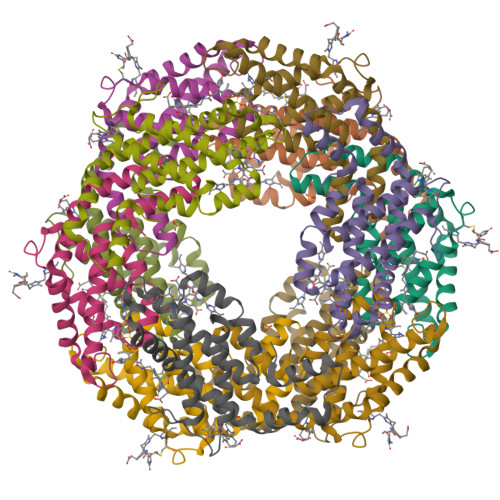
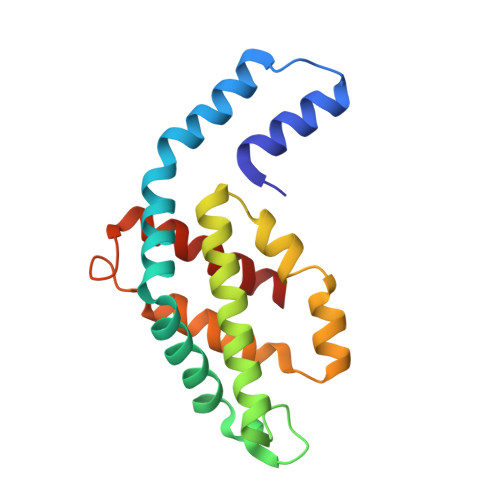
Here, an unprecedented biorefinery approach has been designed to recover high-added value bioproducts starting from the culture of Porphyridium cruentum . This unicellular marine red alga can secrete and accumulate high-value compounds that can find applications in a wide variety of industrial fields. 300 ± 67 mg/L of exopolysaccharides were obtained from cell culture medium; phycoerythrin was efficiently extracted (40% of total extract) and isolated by single chromatography, with a purity grade that allowed the crystal structure determination at 1.60 Å; a twofold increase in β-carotene yield was obtained from the residual biomass; the final residual biomass was found to be enriched in saturated fatty acids. Thus, for the first time, a complete exploitation of P. cruentum culture was set up.
Organizational Affiliation:
Department of Chemical Sciences, University of Naples Federico II, via Cinthia 4, Naples80126, Italy.