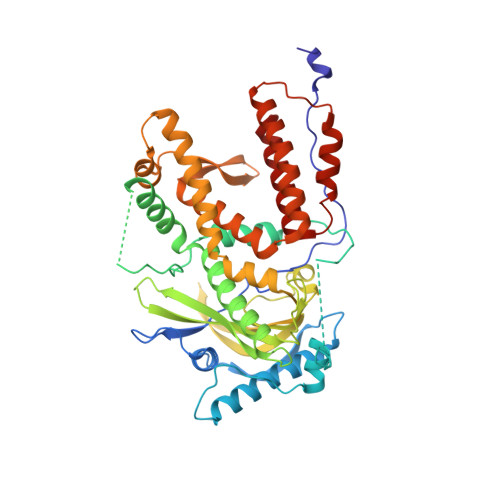
Thermodynamic and structural characterization of an optimized peptide-based inhibitor of the influenza polymerase PA-PB1 subunit interaction.
Radilova, K., Zima, V., Kral, M., Machara, A., Majer, P., Hodek, J., Weber, J., Brynda, J., Strmen, T., Konvalinka, J., Kozisek, M.(2022) Antiviral Res 208: 105449-105449
- PubMed: 36265804
- DOI: https://doi.org/10.1016/j.antiviral.2022.105449
- Primary Citation of Related Structures:
7ZPY - PubMed Abstract:
Influenza virus causes severe respiratory infection in humans. Current antivirotics target three key proteins in the viral life cycle: neuraminidase, the M2 channel and the endonuclease domain of RNA-dependent-RNA polymerase. Due to the development of novel pandemic strains, additional antiviral drugs targetting different viral proteins are still needed. The protein-protein interaction between polymerase subunits PA and PB1 is one such possible target. We recently identified a modified decapeptide derived from the N-terminus of the PB1 subunit with high affinity for the C-terminal part of the PA subunit. Here, we optimized its amino acid hotspots to maintain the inhibitory potency and greatly increase peptide solubility. This allowed thermodynamic characterization of peptide binding to PA. Solving the X-ray structure of the peptide-PA complex provided structural insights into the interaction. Additionally, we optimized intracellular delivery of the peptide using a bicyclic strategy that led to improved inhibition in cell-based assays.
Organizational Affiliation:
Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences, Flemingovo n. 2, 16610, Prague 6, Czech Republic; First Faculty of Medicine, Charles University, Kateřinská 1660/32, 12108, Prague, 2, Czech Republic.