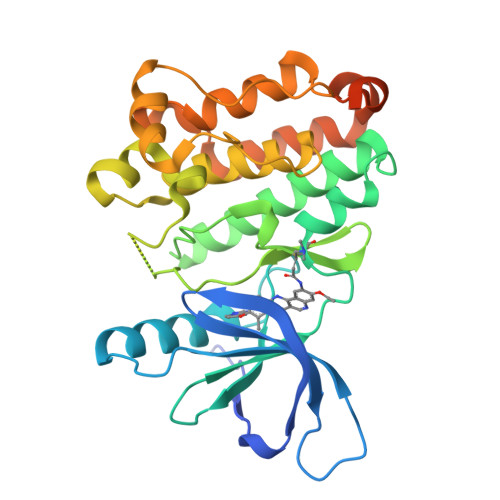
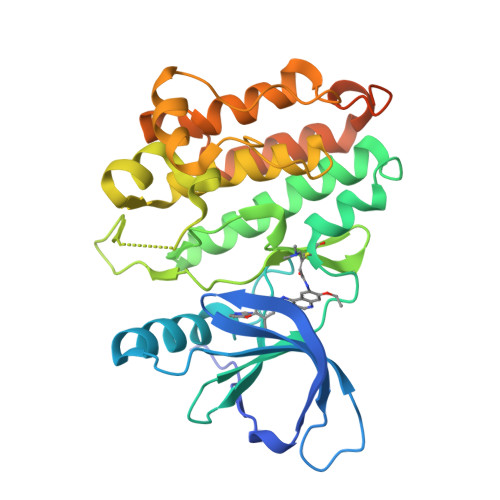
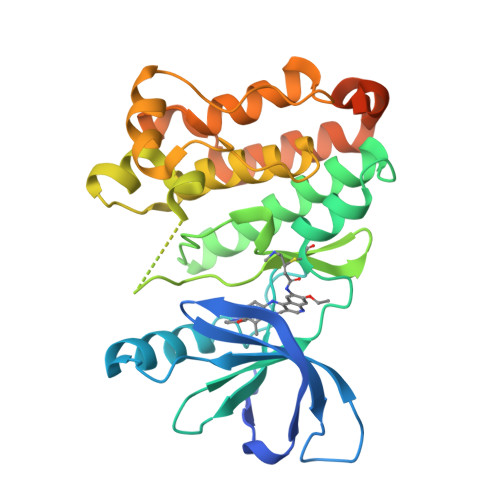
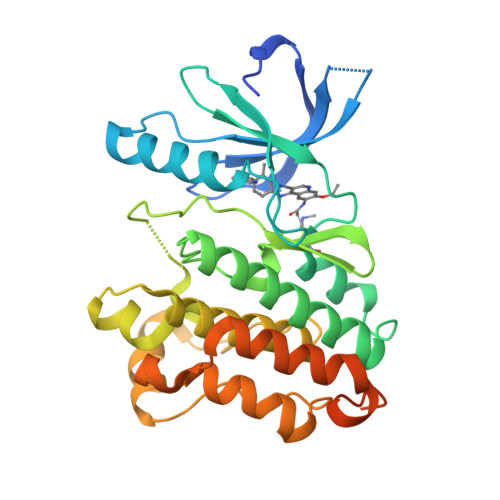
A Novel HER2-Selective Kinase Inhibitor Is Effective in HER2 Mutant and Amplified Non-Small Cell Lung Cancer.
Son, J., Jang, J., Beyett, T.S., Eum, Y., Haikala, H.M., Verano, A., Lin, M., Hatcher, J.M., Kwiatkowski, N.P., Eser, P.O., Poitras, M.J., Wang, S., Xu, M., Gokhale, P.C., Cameron, M.D., Eck, M.J., Gray, N.S., Janne, P.A.(2022) Cancer Res 82: 1633-1645
- PubMed: 35149586
- DOI: https://doi.org/10.1158/0008-5472.CAN-21-2693
- Primary Citation of Related Structures:
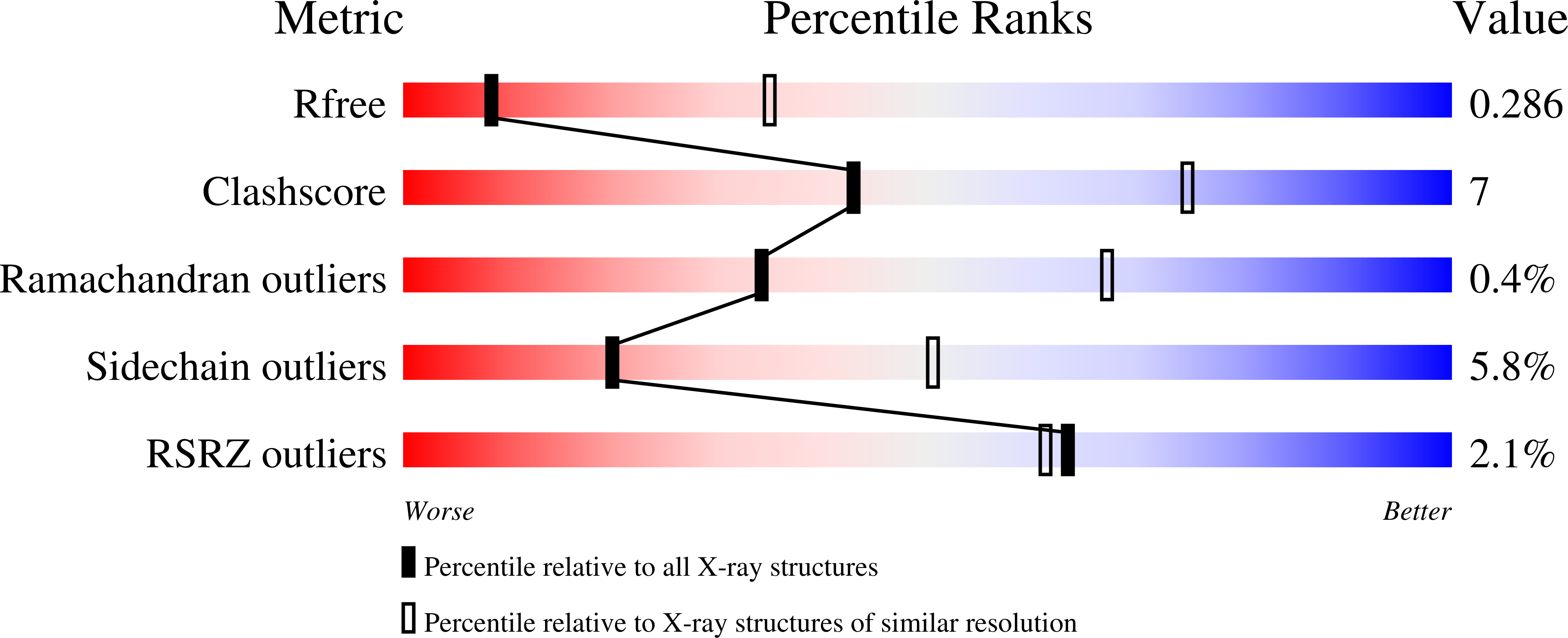
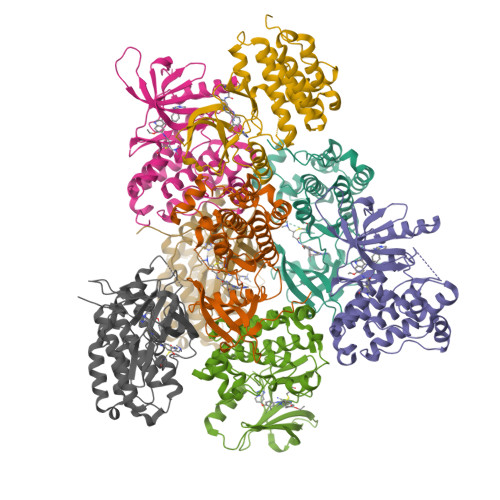
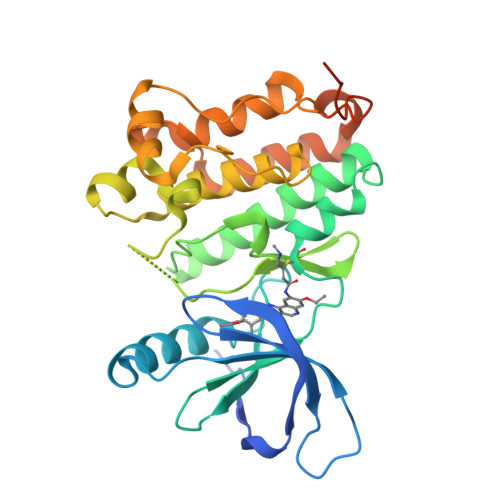
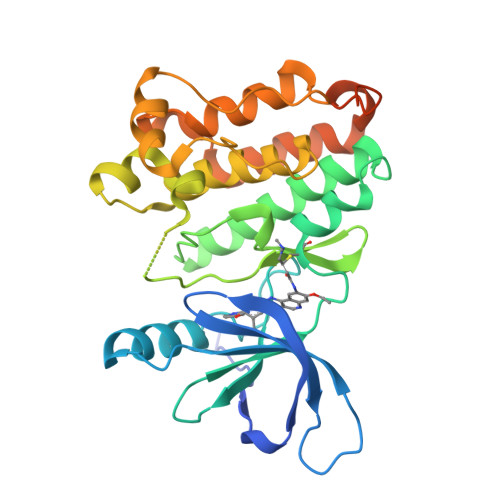
7JXH - PubMed Abstract:
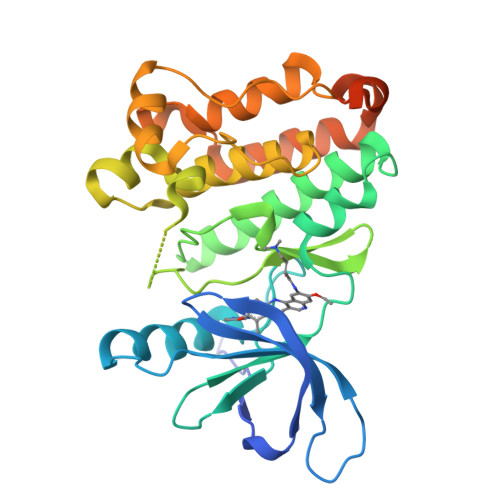
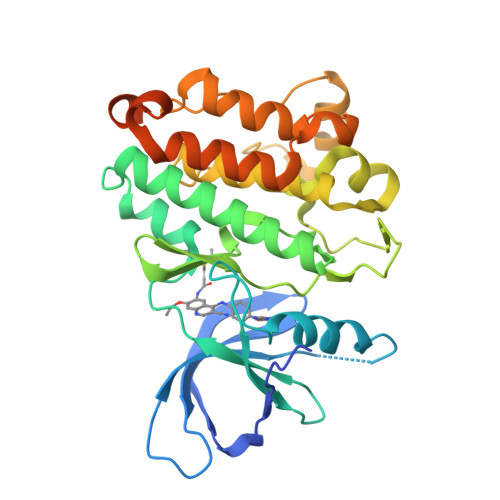
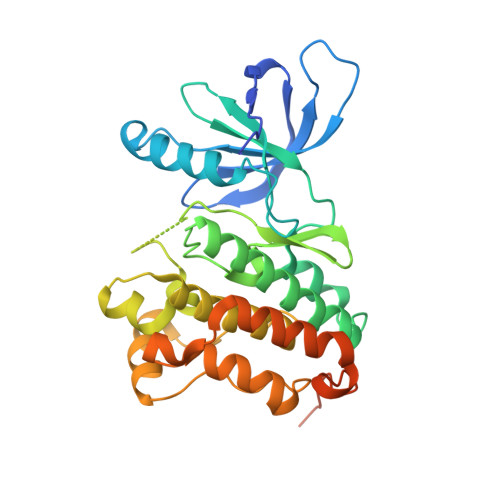
In-frame insertions in exon 20 of HER2 are the most common HER2 mutations in patients with non-small cell lung cancer (NSCLC), a disease in which approved EGFR/HER2 tyrosine kinase inhibitors (TKI) display poor efficiency and undesirable side effects due to their strong inhibition of wild-type (WT) EGFR. Here, we report a HER2-selective covalent TKI, JBJ-08-178-01, that targets multiple HER2 activating mutations, including exon 20 insertions as well as amplification. JBJ-08-178-01 displayed strong selectivity toward HER2 mutants over WT EGFR compared with other EGFR/HER2 TKIs. Determination of the crystal structure of HER2 in complex with JBJ-08-178-01 suggests that an interaction between the inhibitor and Ser783 may be responsible for HER2 selectivity. The compound showed strong antitumoral activity in HER2-mutant or amplified cancers in vitro and in vivo. Treatment with JBJ-08-178-01 also led to a reduction in total HER2 by promoting proteasomal degradation of the receptor. Taken together, the dual activity of JBJ-08-178-01 as a selective inhibitor and destabilizer of HER2 represents a combination that may lead to better efficacy and tolerance in patients with NSCLC harboring HER2 genetic alterations or amplification. This study describes unique mechanisms of action of a new mutant-selective HER2 kinase inhibitor that reduces both kinase activity and protein levels of HER2 in lung cancer.
Organizational Affiliation:
Lowe Center for Thoracic Oncology, Dana-Farber Cancer Institute, Boston, Massachusetts.