Selenols: a new class of carbonic anhydrase inhibitors.
Angeli, A., Tanini, D., Nocentini, A., Capperucci, A., Ferraroni, M., Gratteri, P., Supuran, C.T.(2019) Chem Commun (Camb) 55: 648-651
- PubMed: 30560259
- DOI: https://doi.org/10.1039/c8cc08562e
- Primary Citation of Related Structures:
6HWZ, 6HX5, 6SWM - PubMed Abstract:
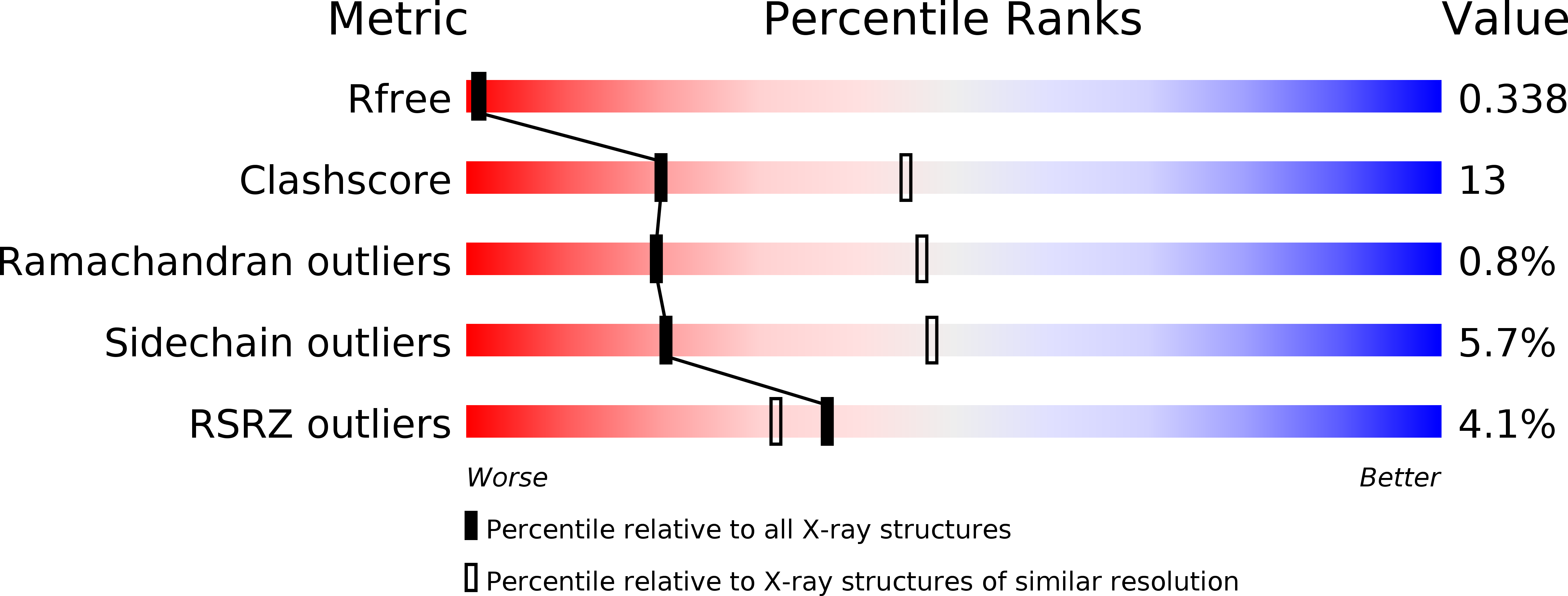
Stable aryl selenols were obtained through a convenient procedure. Selenols represent a new chemotype acting as carbonic anhydrase inhibitors (CAIs), inhibiting four human isoforms, CAs I, II, VII and the tumor-associated CA IX. X-ray crystallographic, physical and computational studies provided insights into the binding mode of this conceptually new class of CAIs.
Organizational Affiliation:
University of Florence, NEUROFARBA Department, Sezione di Scienze Farmaceutiche, Via Ugo Schiff 6, 50019 Sesto Fiorentino, Florence, Italy. claudiu.supuran@unifi.it.