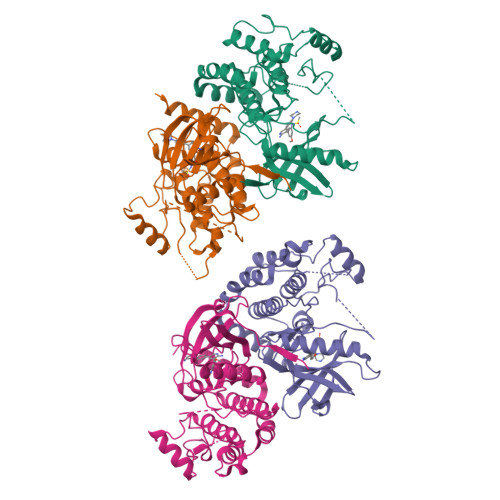
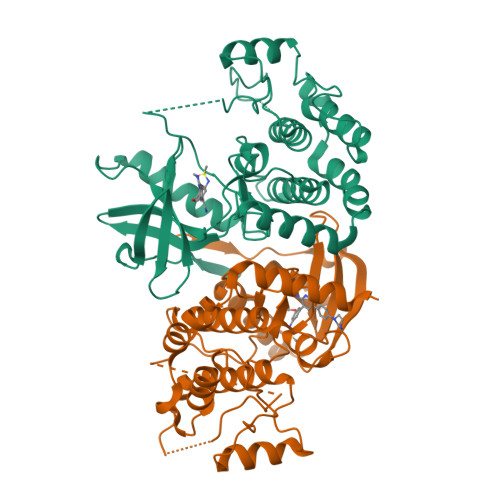
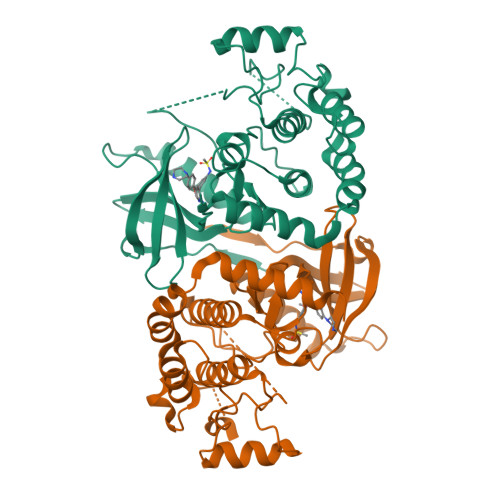
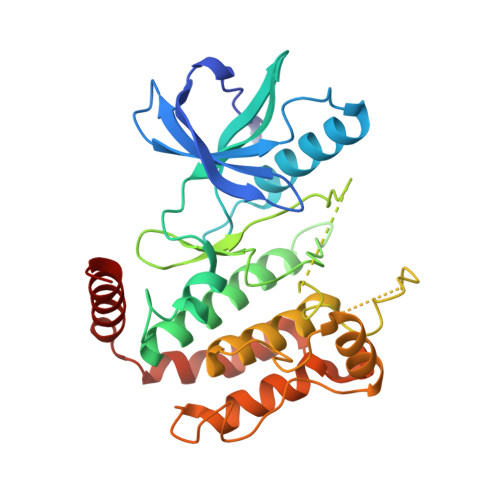
Design of 3,5-diaryl-2-aminopyridines as receptor-interacting protein kinase 2 (RIPK2) and nucleotide-binding oligomerization domain (NOD) cell signaling inhibitors
Suebsuwong, C., Dai, B., Pinkas, D.M., Bufton, J.C., Duddupudi, A.L., Li, L., Hrdinka, M., Schlicher, L., Gyrd-Hansen, M., Hu, M., Bullock, A.N., Degterev, A., Cuny, G.D.To be published.