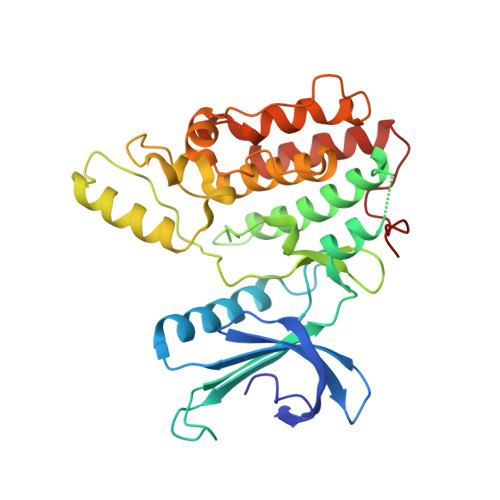
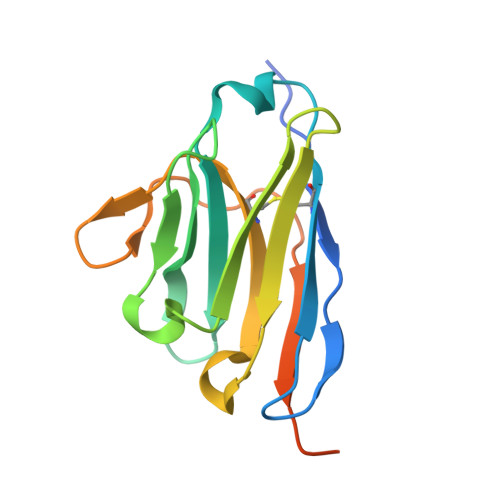
Structural Basis for the Inhibition of Cyclin G-Associated Kinase by Gefitinib.
Ohbayashi, N., Murayama, K., Kato-Murayama, M., Kukimoto-Niino, M., Uejima, T., Matsuda, T., Ohsawa, N., Yokoyoma, S., Nojima, H., Shirouzu, M.(2018) ChemistryOpen 7: 721-727
- PubMed: 30214852
- DOI: https://doi.org/10.1002/open.201800177
- Primary Citation of Related Structures:
5Y7Z, 5Y80 - PubMed Abstract:
Gefitinib is the molecular target drug for advanced non-small-cell lung cancer. The primary target of gefitinib is the positive mutation of epidermal growth factor receptor, but it also inhibits cyclin G-associated kinase (GAK). To reveal the molecular bases of GAK and gefitinib binding, structure analyses were conducted and determined two forms of the gefitinib-bound nanobody⋅GAK kinase domain complex structures. The first form, GAK_1, has one gefitinib at the ATP binding pocket, whereas the second form, GAK_2, binds one each in the ATP binding site and a novel binding site adjacent to the activation segment C-terminal helix, a unique element of the Numb-associated kinase family. In the novel binding site, gefitinib binds in the hydrophobic groove around the activation segment, disrupting the conserved hydrogen bonds for the catalytic activity. These structures suggest possibilities for the development of selective GAK inhibitors for viral infections, such as the hepatitis C virus.
Organizational Affiliation:
Division of Structural and Synthetic Biology RIKEN Center for Life Science Technologies 1-7-22 Suehiro-cho, Tsurumi Yokohama 230-0045 Japan.