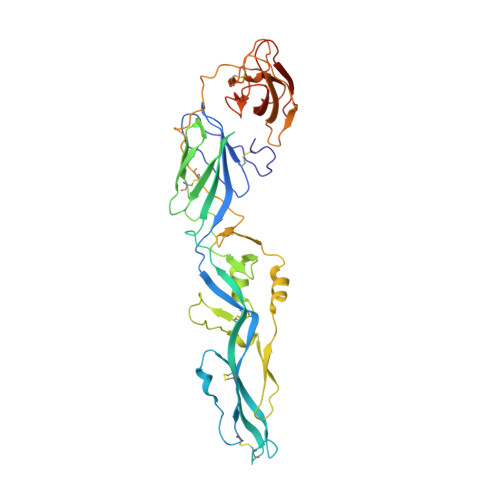
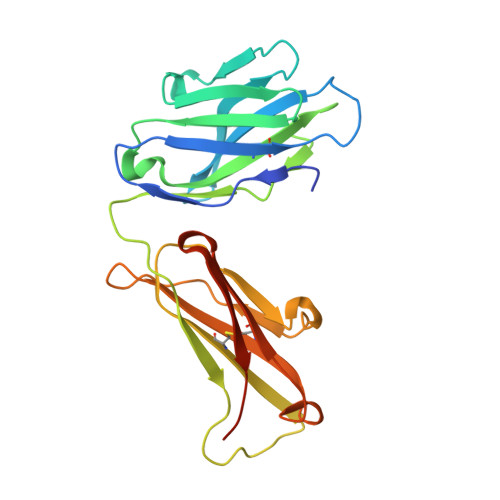
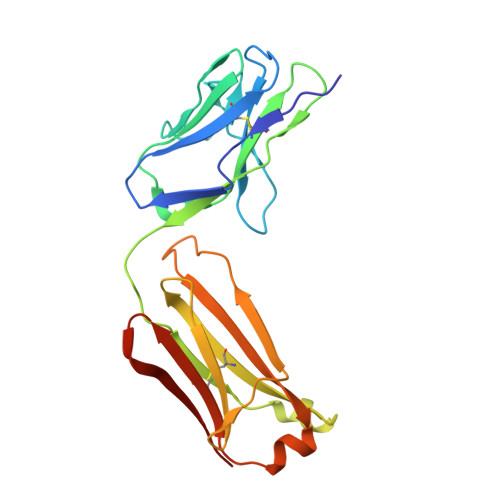
Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody.
Dai, L., Song, J., Lu, X., Deng, Y.Q., Musyoki, A.M., Cheng, H., Zhang, Y., Yuan, Y., Song, H., Haywood, J., Xiao, H., Yan, J., Shi, Y., Qin, C.F., Qi, J., Gao, G.F.(2016) Cell Host Microbe 19: 696-704
- PubMed: 27158114
- DOI: https://doi.org/10.1016/j.chom.2016.04.013
- Primary Citation of Related Structures:
5JHL, 5JHM - PubMed Abstract:
Zika virus (ZIKV), a mosquito-borne flavivirus, is a current global public health concern. The flavivirus envelope (E) glycoprotein is responsible for virus entry and represents a major target of neutralizing antibodies for other flaviviruses. Here, we report the structures of ZIKV E protein at 2.0 Å and in complex with a flavivirus broadly neutralizing murine antibody 2A10G6 at 3.0 Å. ZIKV-E resembles all the known flavivirus E structures but contains a unique, positively charged patch adjacent to the fusion loop region of the juxtaposed monomer, which may influence host attachment. The ZIKV-E-2A10G6 complex structure reveals antibody recognition of a highly conserved fusion loop. 2A10G6 binds to ZIKV-E with high affinity in vitro and neutralizes currently circulating ZIKV strains in vitro and in mice. The E protein fusion loop epitope represents a potential candidate for therapeutic antibodies against ZIKV.
Organizational Affiliation:
Research Network of Immunity and Health (RNIH), Beijing Institutes of Life Science, Chinese Academy of Sciences, Beijing 100101, China.