PI4KB in complex with Rab11 and the MI359 Inhibitor
Chalupska, D., Mejdrova, I., Nencka, R., Boura, E.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | 572 | Homo sapiens | Mutation(s): 0 Gene Names: PI4KB, PIK4CB EC: 2.7.1.67 | 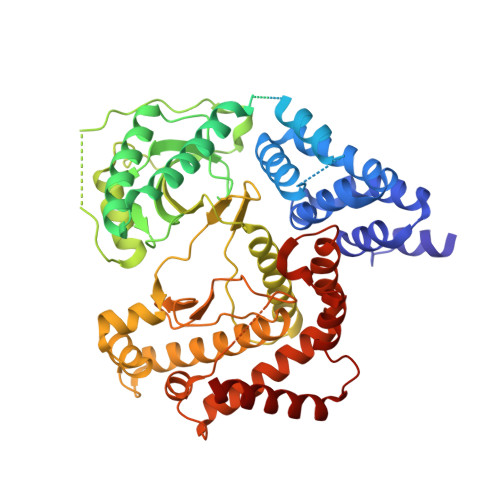 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9UBF8 (Homo sapiens) Explore Q9UBF8 Go to UniProtKB: Q9UBF8 | |||||
PHAROS: Q9UBF8 GTEx: ENSG00000143393 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9UBF8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ras-related protein Rab-11A | 221 | Homo sapiens | Mutation(s): 1 Gene Names: RAB11A, RAB11 EC: 3.6.5.2 | 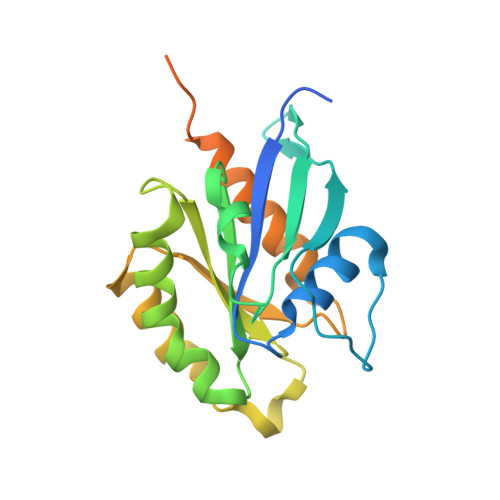 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62491 (Homo sapiens) Explore P62491 Go to UniProtKB: P62491 | |||||
PHAROS: P62491 GTEx: ENSG00000103769 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62491 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 5W7 Query on 5W7 | C [auth A] | ~{N}-[2-[[3-[3-[(4-azanylcyclohexyl)sulfamoyl]-4-methoxy-phenyl]-6-chloranyl-2-methyl-imidazo[1,2-b]pyridazin-8-yl]amino]ethyl]ethanamide C24 H32 Cl N7 O4 S HORACVGDHNAIMC-IYARVYRRSA-N |  | ||
| GSP Query on GSP | D [auth B] | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE C10 H16 N5 O13 P3 S XOFLBQFBSOEHOG-UUOKFMHZSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 49.266 | α = 90 |
| b = 103.937 | β = 90 |
| c = 187.646 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |