Development of a Potent, Specific CDK8 Kinase Inhibitor Which Phenocopies CDK8/19 Knockout Cells.
Koehler, M.F., Bergeron, P., Blackwood, E.M., Bowman, K., Clark, K.R., Firestein, R., Kiefer, J.R., Maskos, K., McCleland, M.L., Orren, L., Salphati, L., Schmidt, S., Schneider, E.V., Wu, J., Beresini, M.H.(2016) ACS Med Chem Lett 7: 223-228
- PubMed: 26985305
- DOI: https://doi.org/10.1021/acsmedchemlett.5b00278
- Primary Citation of Related Structures:
5CEI - PubMed Abstract:
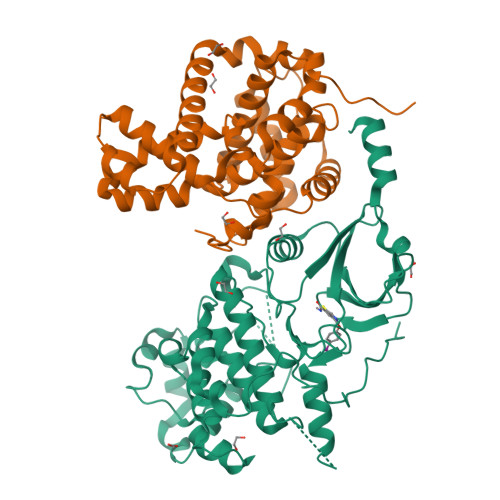
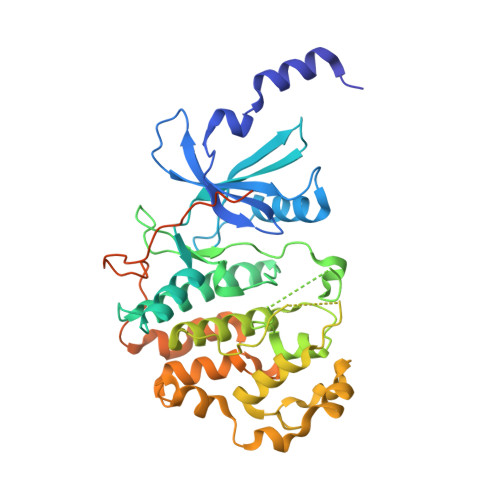
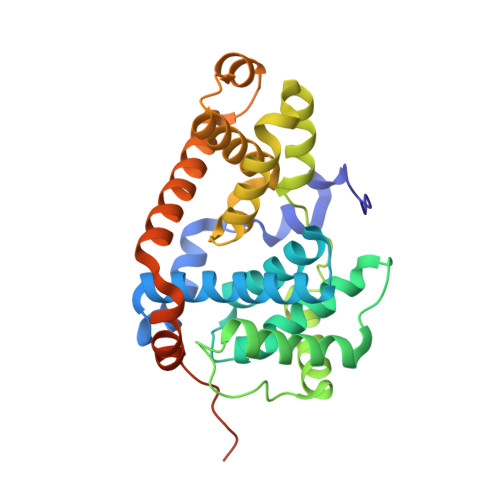
Beginning with promiscuous COT inhibitors, which were found to inhibit CDK8, a series of 6-aza-benzothiophene containing compounds were developed into potent, selective CDK8 inhibitors. When cocrystallized with CDK8 and cyclin C, these compounds exhibit an unusual binding mode, making a single hydrogen bond to the hinge residue A100, a second to K252, and a key cation-π interaction with R356. Structure-based drug design resulted in tool compounds 13 and 32, which are highly potent, kinase selective, permeable compounds with a free fraction >2% and no measurable efflux. Despite these attractive properties, these compounds exhibit weak antiproliferative activity in the HCT-116 colon cancer cell line. Further examination of the activity of 32 in this cell line revealed that the compound reduced phosphorylation of the known CDK8 substrate STAT1 in a manner identical to a CDK8 knockout clone, illustrating the complex effects of inhibition of CDK8 kinase activity in proliferation in these cells.
Organizational Affiliation:
Departments of Discovery Chemistry, Translational Oncology, Structural Biology, Biochemical and Cellular Pharmacology, Pathology, Drug Metabolism and Pharmacokinetics, and Structural Biology, Genentech, Inc. , 1 DNA Way, South San Francisco, California 94080, United States.