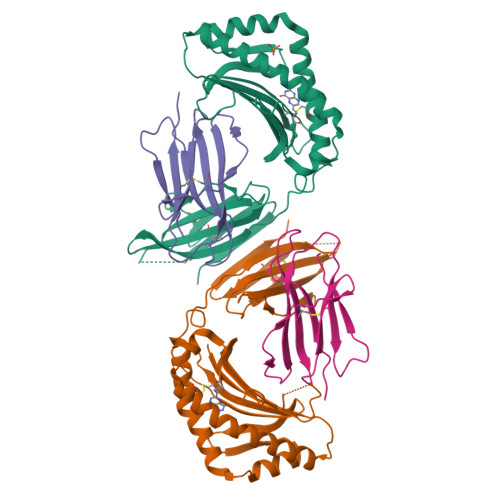
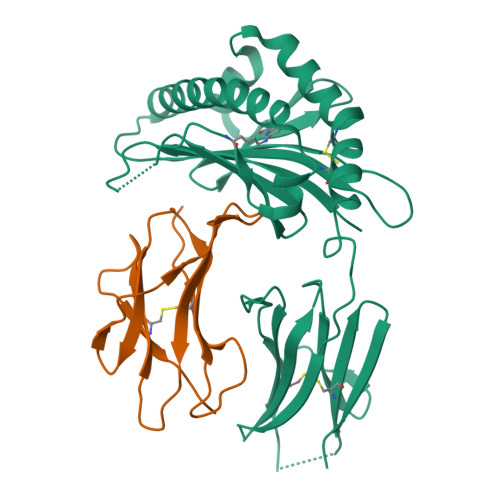
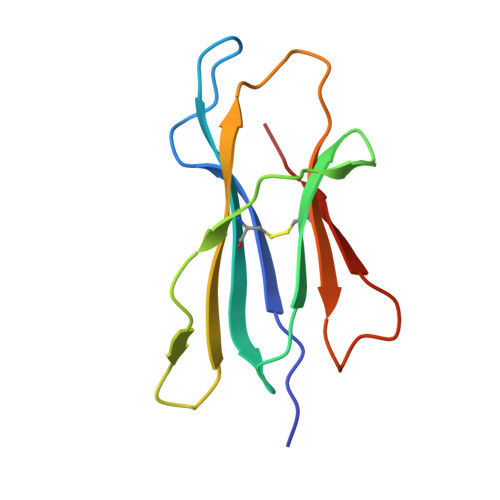
MR1 presents microbial vitamin B metabolites to MAIT cells
Kjer-Nielsen, L., Patel, O., Corbett, A.J., Le Nours, J., Meehan, B., Liu, L., Bhati, M., Chen, Z., Kostenko, L., Reantragoon, R., Williamson, N.A., Purcell, A.W., Dudek, N.L., McConville, M.J., O'Hair, R.A.J., Khairallah, G.N., Godfrey, D.I., Fairlie, D.P., Rossjohn, J., McCluskey, J.(2012) Nature 491: 717-723
- PubMed: 23051753
- DOI: https://doi.org/10.1038/nature11605
- Primary Citation of Related Structures:
4GUP - PubMed Abstract:
Antigen-presenting molecules, encoded by the major histocompatibility complex (MHC) and CD1 family, bind peptide- and lipid-based antigens, respectively, for recognition by T cells. Mucosal-associated invariant T (MAIT) cells are an abundant population of innate-like T cells in humans that are activated by an antigen(s) bound to the MHC class I-like molecule MR1. Although the identity of MR1-restricted antigen(s) is unknown, it is present in numerous bacteria and yeast. Here we show that the structure and chemistry within the antigen-binding cleft of MR1 is distinct from the MHC and CD1 families. MR1 is ideally suited to bind ligands originating from vitamin metabolites. The structure of MR1 in complex with 6-formyl pterin, a folic acid (vitamin B9) metabolite, shows the pterin ring sequestered within MR1. Furthermore, we characterize related MR1-restricted vitamin derivatives, originating from the bacterial riboflavin (vitamin B2) biosynthetic pathway, which specifically and potently activate MAIT cells. Accordingly, we show that metabolites of vitamin B represent a class of antigen that are presented by MR1 for MAIT-cell immunosurveillance. As many vitamin biosynthetic pathways are unique to bacteria and yeast, our data suggest that MAIT cells use these metabolites to detect microbial infection.
Organizational Affiliation:
Department of Microbiology & Immunology, University of Melbourne, Parkville, Victoria 3010, Australia.