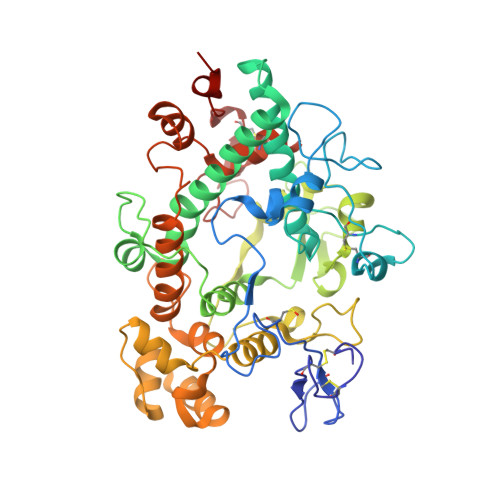
Potent Reversible Inhibition of Myeloperoxidase by Aromatic Hydroxamates
Forbes, L.V., Sjogren, T., Auchere, F., Jenkins, D.W., Thong, B., Laughton, D., Hemsley, P., Pairaudeau, G., Turner, R., Eriksson, H., Unitt, J.F., Kettle, A.J.(2013) J Biol Chem 288: 36636
- PubMed: 24194519
- DOI: https://doi.org/10.1074/jbc.M113.507756
- Primary Citation of Related Structures:
4C1M - PubMed Abstract:
The neutrophil enzyme myeloperoxidase (MPO) promotes oxidative stress in numerous inflammatory pathologies by producing hypohalous acids. Its inadvertent activity is a prime target for pharmacological control. Previously, salicylhydroxamic acid was reported to be a weak reversible inhibitor of MPO. We aimed to identify related hydroxamates that are good inhibitors of the enzyme. We report on three hydroxamates as the first potent reversible inhibitors of MPO. The chlorination activity of purified MPO was inhibited by 50% by a 5 nm concentration of a trifluoromethyl-substituted aromatic hydroxamate, HX1. The hydroxamates were specific for MPO in neutrophils and more potent toward MPO compared with a broad range of redox enzymes and alternative targets. Surface plasmon resonance measurements showed that the strength of binding of hydroxamates to MPO correlated with the degree of enzyme inhibition. The crystal structure of MPO-HX1 revealed that the inhibitor was bound within the active site cavity above the heme and blocked the substrate channel. HX1 was a mixed-type inhibitor of the halogenation activity of MPO with respect to both hydrogen peroxide and halide. Spectral analyses demonstrated that hydroxamates can act variably as substrates for MPO and convert the enzyme to a nitrosyl ferrous intermediate. This property was unrelated to their ability to inhibit MPO. We propose that aromatic hydroxamates bind tightly to the active site of MPO and prevent it from producing hypohalous acids. This mode of reversible inhibition has potential for blocking the activity of MPO and limiting oxidative stress during inflammation.
Organizational Affiliation:
From the Centre for Free Radical Research, Department of Pathology, University of Otago Christchurch, Christchurch 8140, New Zealand.