X-Ray Structural Analysis of Tau-Tubulin Kinase 1 and its Interactions with Small Molecular Inhibitors.
Xue, Y., Wan, P.T., Hillertz, P., Schweikart, F., Zhao, Y., Wissler, L., Dekker, N.(2013) ChemMedChem 8: 1846
- PubMed: 24039150
- DOI: https://doi.org/10.1002/cmdc.201300274
- Primary Citation of Related Structures:
4BTJ, 4BTK, 4BTM - PubMed Abstract:
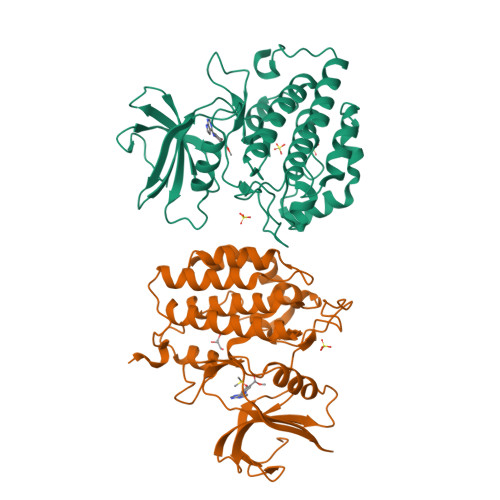
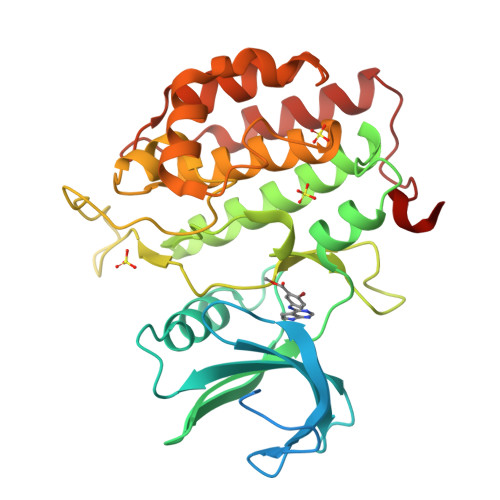
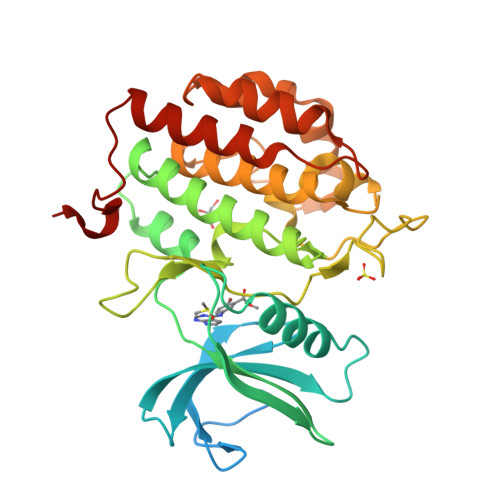
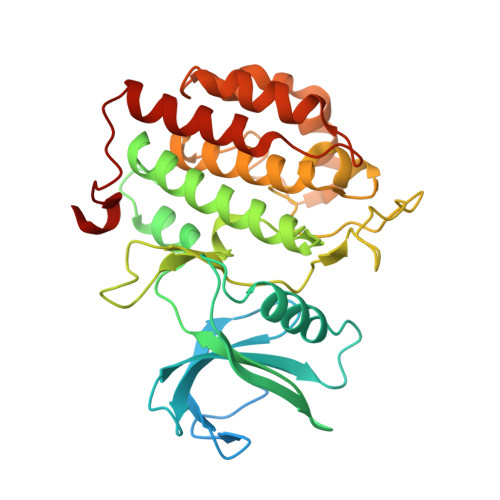
Tau-tubulin kinase 1 (TTBK1) is a serine/threonine/tyrosine kinase that putatively phosphorylates residues including S422 in tau protein. Hyperphosphorylation of tau protein is the primary cause of tau pathology and neuronal death associated with Alzheimer's disease. A library of 12 truncation variants comprising the TTBK1 kinase domain was screened for expression in Escherichia coli and insect cells. One variant (residues 14-313) could be purified, but mass spectrometric analysis revealed extensive phosphorylation of the protein. Co-expression with lambda phosphatase in E. coli resulted in production of homogeneous, nonphosphorylated TTBK1. Binding of ATP and several compounds to TTBK1 was characterized by surface plasmon resonance. Crystal structures of TTBK1 in the unliganded form and in complex with ATP, and two high-affinity ATP-competitive inhibitors, 3-[(6,7-dimethoxyquinazolin-4-yl)amino]phenol (1) and methyl 2-bromo-5-(7H-pyrrolo[2,3-d]pyrimidin-4-ylamino)benzoate (2), were elucidated. The structure revealed two clear basic patches near the ATP pocket providing an explanation of TTBK1 for phosphorylation-primed substrates. Interestingly, compound 2 displayed slow binding kinetics to TTBK1, the structure of TTBK1 in complex with this compound revealed a reorganization of the L199-D200 peptide backbone conformation together with altered hydrogen bonding with compound 2. These conformational changes necessary for the binding of compound 2 are likely the basis of the slow kinetics. This first TTBK1 structure can assist the discovery of novel inhibitors for the treatment of Alzheimer's disease.
Organizational Affiliation:
Discovery Sciences, AstraZeneca R&D Mölndal, 43183 Mölndal (Sweden). yafeng.xue@astrazeneca.com.