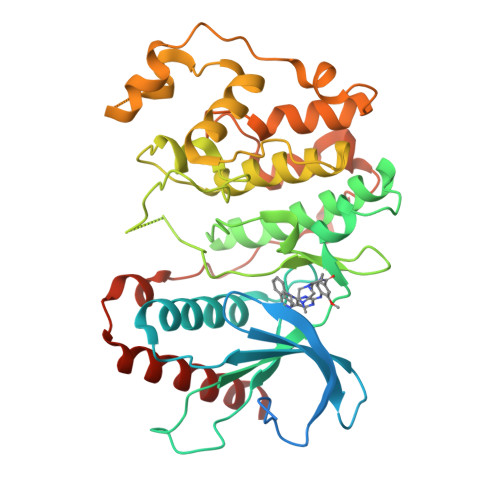
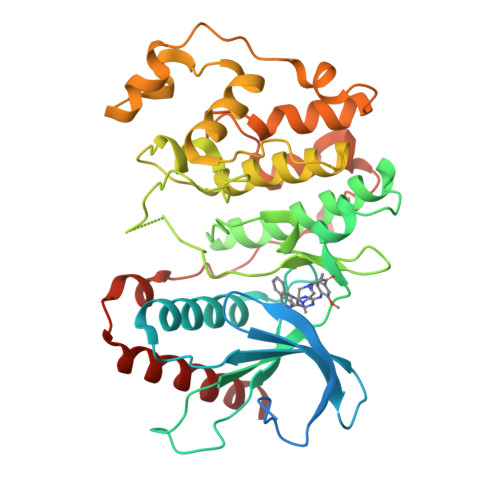
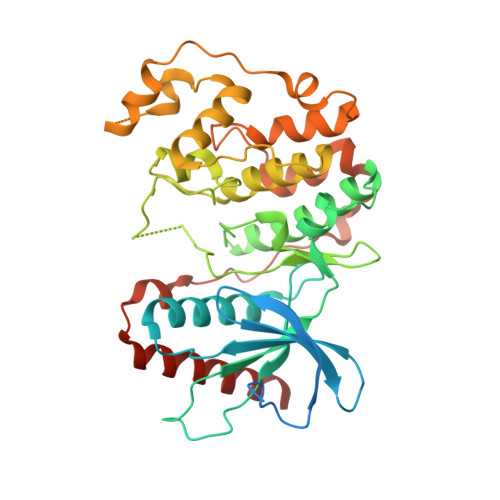
X-Ray Crystal Structure of Erk5 (Mapk7) in Complex with a Specific Inhibitor.
Elkins, J.M., Wang, J., Deng, X., Pattison, M.J., Arthur, S., Erazo, T., Gomez, N., Lizcano, J.M., Gray, N.S., Knapp, S.(2013) J Med Chem 56: 4413
- PubMed: 23656407
- DOI: https://doi.org/10.1021/jm4000837
- Primary Citation of Related Structures:
4B99 - PubMed Abstract:
The protein kinase ERK5 (MAPK7) is an emerging drug target for a variety of indications, in particular for cancer where it plays a key role mediating cell proliferation, survival, epithelial-mesenchymal transition, and angiogenesis. To date, no three-dimensional structure has been published that would allow rational design of inhibitors. To address this, we determined the X-ray crystal structure of the human ERK5 kinase domain in complex with a highly specific benzo[e]pyrimido[5,4-b]diazepine-6(11H)-one inhibitor. The structure reveals that specific residue differences in the ATP-binding site, compared to the related ERKs p38s and JNKs, allow for the development of ERK5-specific inhibitors. The selectivity of previously observed ERK5 inhibitors can also be rationalized using this structure, which provides a template for future development of inhibitors with potential for treatment of disease.
Organizational Affiliation:
Structural Genomics Consortium, Nuffield Department of Clinical Medicine, University of Oxford, Old Road Campus Research Building, Roosevelt Drive, Oxford, OX3 7DQ, UK.