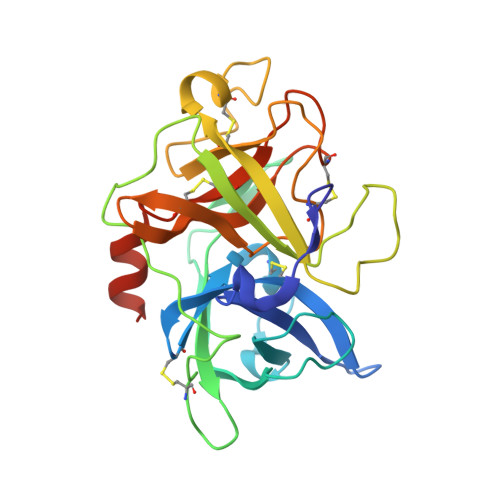
Bicyclic peptide inhibitor reveals large contact interface with a protease target
Angelini, A., Cendron, L., Chen, S., Touati, J., Winter, G., Zanotti, G., Heinis, C.(2012) ACS Chem Biol 7: 817-821
- PubMed: 22304751
- DOI: https://doi.org/10.1021/cb200478t
- Primary Citation of Related Structures:
3QN7 - PubMed Abstract:
From a large combinatorial library of chemically constrained bicyclic peptides we isolated a selective and potent (K(i) = 53 nM) inhibitor of human urokinase-type plasminogen activator (uPA) and crystallized the complex. This revealed an extended structure of the peptide with both peptide loops engaging the target to form a large interaction surface of 701 Å(2) with multiple hydrogen bonds and complementary charge interactions, explaining the high affinity and specificity of the inhibitor. The interface resembles that between two proteins and suggests that these constrained peptides have the potential to act as small protein mimics.
Organizational Affiliation:
Institute of Chemical Sciences and Engineering, Ecole Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland.