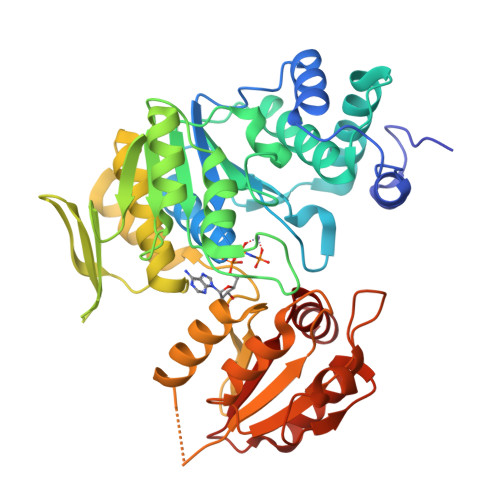
Crystal structure of bifunctional folylpolyglutamate synthase/dihydrofolate synthase complexed with AMPpnp and Mn ion from Yersinia pestis c092
Nocek, B., Makowska-Grzyska, M., Maltseva, N., Anderson, W., Joachimiak, A., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.