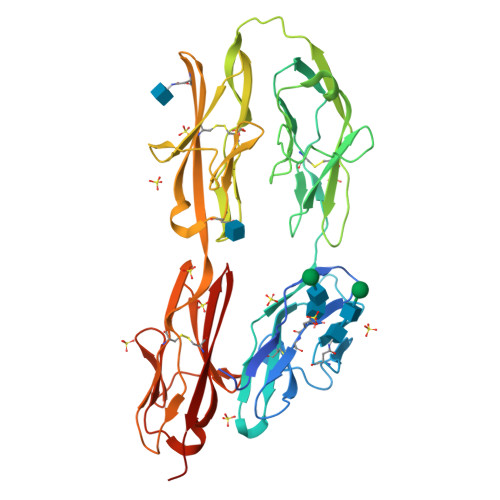
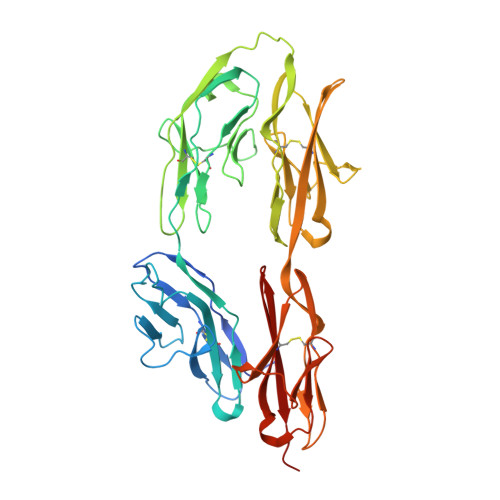
N-terminal horseshoe conformation of DCC is functionally required for axon guidance and might be shared by other neural receptors.
Chen, Q., Sun, X., Zhou, X.H., Liu, J.H., Wu, J., Zhang, Y., Wang, J.H.(2013) J Cell Sci 126: 186-195
- PubMed: 23038776
- DOI: https://doi.org/10.1242/jcs.111278
- Primary Citation of Related Structures:
3LAF - PubMed Abstract:
Deleted in colorectal cancer (DCC) is a receptor for the axon guidance cues netrin-1 and draxin. The interactions between these guidance cues and DCC play a key role in the development of the nervous system. In the present study, we reveal the crystal structure of the N-terminal four Ig-like domains of DCC. The molecule folds into a horseshoe-like configuration. We demonstrate that this horseshoe conformation of DCC is required for guidance-cue-mediated axonal attraction. Structure-based mutations that disrupt the DCC horseshoe indeed impair its function. A comparison of the DCC horseshoe with previously described horseshoe structures has revealed striking conserved structural features and important sequence signatures. Using these signatures, a genome-wide search allows us to predict the N-terminal horseshoe arrangement in a number of other cell surface receptors, nearly all of which function in the nervous system. The N-terminal horseshoe appears to be evolutionally selected as a platform for neural receptors.
Organizational Affiliation:
Department of Oncology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA 02215, USA.