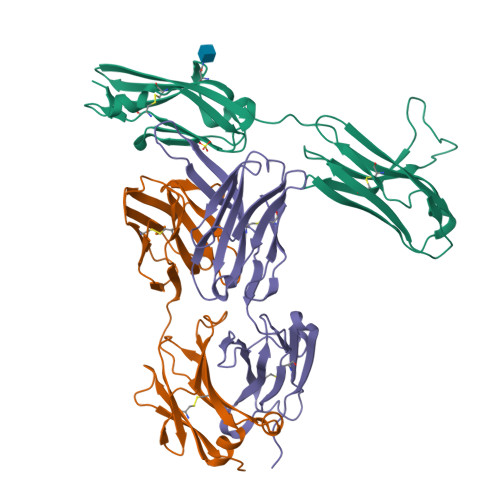
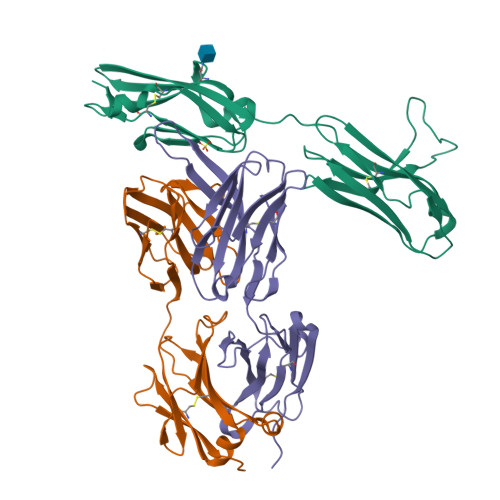
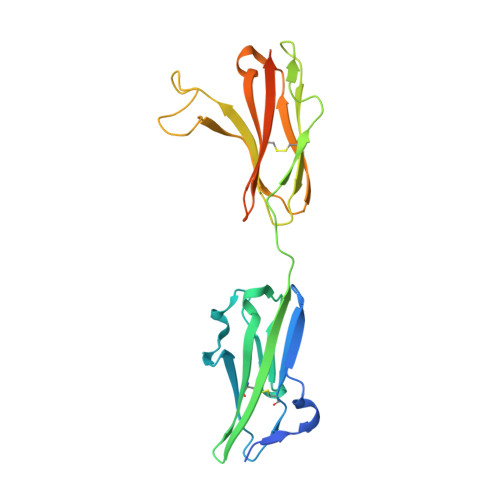
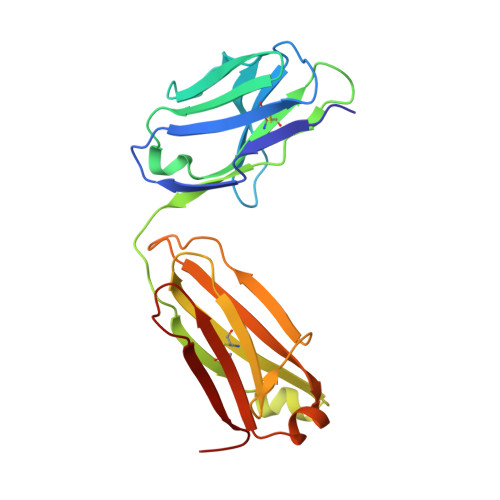
Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice.
Qing, J., Du, X., Chen, Y., Chan, P., Li, H., Wu, P., Marsters, S., Stawicki, S., Tien, J., Totpal, K., Ross, S., Stinson, S., Dornan, D., French, D., Wang, Q.R., Stephan, J.P., Wu, Y., Wiesmann, C., Ashkenazi, A.(2009) J Clin Invest 119: 1216-1229
- PubMed: 19381019
- DOI: https://doi.org/10.1172/JCI38017
- Primary Citation of Related Structures:
3GRW - PubMed Abstract:
Overexpression of FGF receptor 3 (FGFR3) is implicated in the development of t(4;14)-positive multiple myeloma. While FGFR3 is frequently overexpressed and/or activated through mutations in bladder cancer, the functional importance of FGFR3 and its potential as a specific therapeutic target in this disease have not been elucidated in vivo. Here we report that inducible knockdown of FGFR3 in human bladder carcinoma cells arrested cell-cycle progression in culture and markedly attenuated tumor progression in xenografted mice. Further, we developed a unique antibody (R3Mab) that inhibited not only WT FGFR3, but also various mutants of the receptor, including disulfide-linked cysteine mutants. Biochemical analysis and 2.1-A resolution crystallography revealed that R3Mab bound to a specific FGFR3 epitope that simultaneously blocked ligand binding, prevented receptor dimerization, and induced substantial conformational changes in the receptor. R3Mab exerted potent antitumor activity against bladder carcinoma and t(4;14)-positive multiple myeloma xenografts in mice by antagonizing FGFR3 signaling and eliciting antibody-dependent cell-mediated cytotoxicity (ADCC). These studies provide in vivo evidence demonstrating an oncogenic role of FGFR3 in bladder cancer and support antibody-based targeting of FGFR3 in hematologic and epithelial cancers driven by WT or mutant FGFR3.
Organizational Affiliation:
Department of Molecular Oncology, Genentech Inc., South San Francisco, California, USA. jqing@gene.com