Structure of the Aliphatic Sulfonate-Binding Protein Ssua from Escherichia Coli
Beale, J., Lee, S.Y., Iwata, S., Beis, K.(2010) Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 391
- PubMed: 20383006
- DOI: https://doi.org/10.1107/S1744309110006226
- Primary Citation of Related Structures:
2X26 - PubMed Abstract:
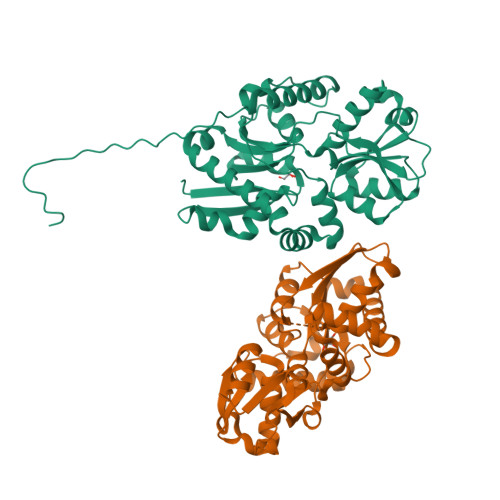
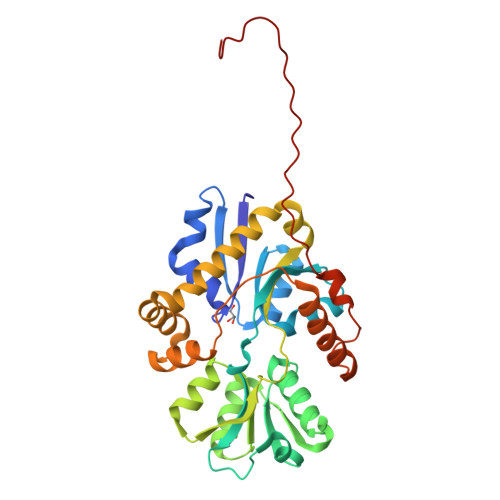
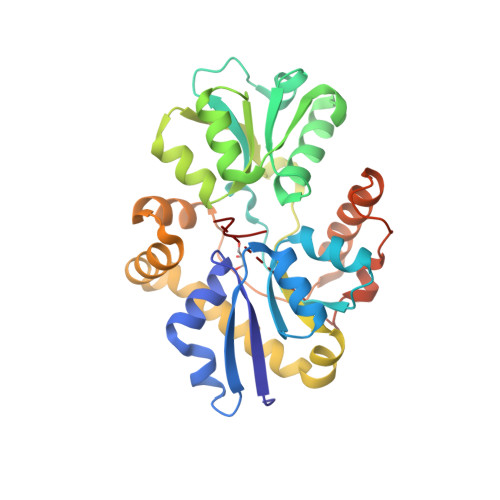
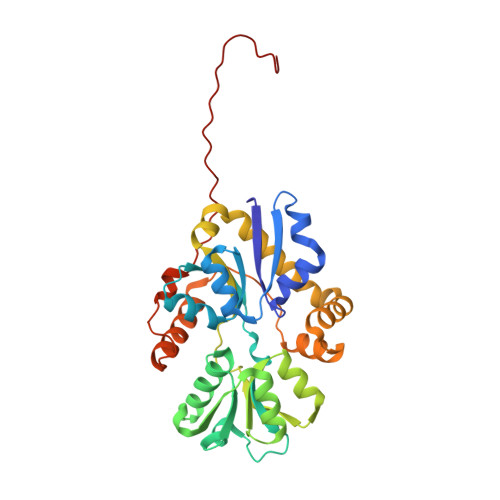
Sulfur is an essential component for the biosynthesis of the sulfur-containing amino acids L-methionine and L-cysteine. Under sulfur-starvation conditions, bacteria are capable of scavenging sulfur from sulfur-containing compounds and transporting it across membranes. Here, the crystal structure of the periplasmic aliphatic sulfonate-binding protein SsuA from Escherichia coli is reported at 1.75 A resolution in the substrate-free state. The overall structure of SsuA resembles the structures of other periplasmic binding proteins and contains two globular domains that form a cleft. Comparison with other periplasmic binding proteins revealed that one of the domains has been displaced by a rigid movement of 17 degrees . Interestingly, the tight crystal packing appears to be mediated by a 13-amino-acid tail from the cloning that folds within the cleft of the next monomer.
Organizational Affiliation:
Membrane Protein Laboratory, Diamond Light Source, Harwell Science and Innovation Campus, Oxfordshire OX11 0DE, England.