X-ray crystallographic and calorimetric studies of the effects of the mutation Trp59-->Tyr in ribonuclease T1.
Schubert, W.D., Schluckebier, G., Backmann, J., Granzin, J., Kisker, C., Choe, H.W., Hahn, U., Pfeil, W., Saenger, W.(1994) Eur J Biochem 220: 527-534
- PubMed: 8125111
- DOI: https://doi.org/10.1111/j.1432-1033.1994.tb18652.x
- Primary Citation of Related Structures:
1TRP, 1TRQ - PubMed Abstract:
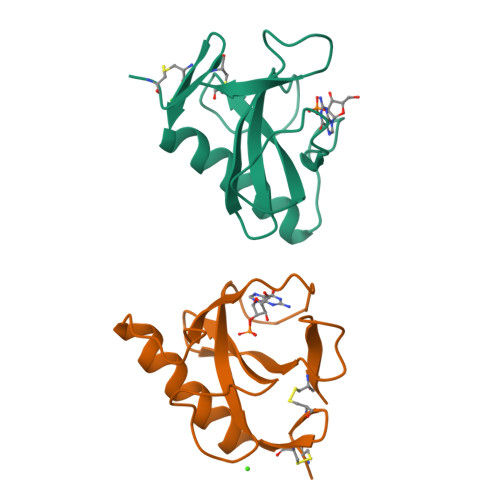
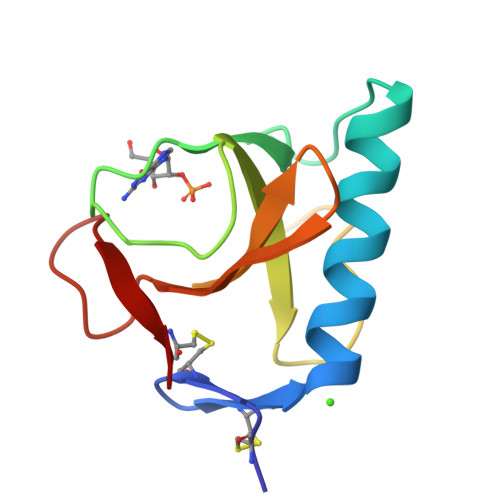
Two mutants of ribonuclease T1 (RNaseT1), [59-tyrosine]ribonuclease T1 (W59Y) and [45-tryptophan,59-tyrosine]ribonuclease T1 (Y45W/W59Y) possess between 150% and 190% wild-type activity. They have been crystallised as complexes of the inhibitor 2'-guanylic acid and analysed by X-ray diffraction at resolutions of 0.23 nm and 0.24 nm, respectively. The space group for both is monoclinic, P2(1), with two molecules/asymmetric unit, W59Y: a = 4.934 nm, b = 4.820 nm, c = 4.025 nm, beta = 90.29 degrees. Y45W/W59Y: a = 4.915 nm, b = 4.815 nm, c = 4.015 nm, beta = 90.35 degrees. Compared to wild-type RNaseT1 in complex with 2'-guanylic acid (2'GMP) both mutant inhibitor complexes indicate that the replacement of Trp59 by Tyr leads to a 0.04-nm inward shift of the single alpha-helix and to significant differences in the active-site geometry, inhibitor conformation and inhibitor binding. Calorimetric studies of a range of mutants [24-tryptophan]ribonuclease T1 (Y24W), [42-tryptophan]ribonuclease T1 (Y42W), [45-tryptophan]ribonuclease T1 (Y45W), [92-alanine]ribonuclease T1 (H92A) and [92-threonine]ribonuclease T1 (H92T) with and without the further mutation Trp59-->Tyr showed that mutant proteins for which Trp59 is replaced by Tyr exhibit slightly decreased thermal stability.
Organizational Affiliation:
Institut für Kristallographie, Freien Universität Berlin, Germany.