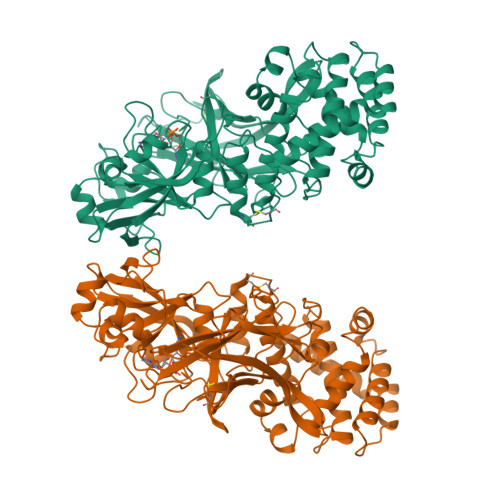
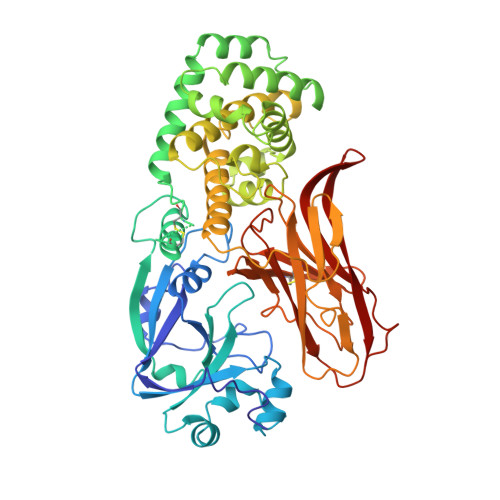
Refined structure of monomeric diphtheria toxin at 2.3 A resolution.
Bennett, M.J., Eisenberg, D.(1994) Protein Sci 3: 1464-1475
- PubMed: 7833808
- DOI: https://doi.org/10.1002/pro.5560030912
- Primary Citation of Related Structures:
1MDT - PubMed Abstract:
The structure of toxic monomeric diphtheria toxin (DT) was determined at 2.3 A resolution by molecular replacement based on the domain structures in dimeric DT and refined to an R factor of 20.7%. The model consists of 2 monomers in the asymmetric unit (1,046 amino acid residues), including 2 bound adenylyl 3'-5' uridine 3' monophosphate molecules and 396 water molecules. The structures of the 3 domains are virtually identical in monomeric and dimeric DT; however, monomeric DT is compact and globular as compared to the "open" monomer within dimeric DT (Bennett MJ, Choe S, Eisenberg D, 1994b, Protein Sci 3:0000-0000). Detailed differences between monomeric and dimeric DT are described, particularly (1) changes in main-chain conformations of 8 residues acting as a hinge to "open" or "close" the receptor-binding (R) domain, and (2) a possible receptor-docking site, a beta-hairpin loop protruding from the R domain containing residues that bind the cell-surface DT receptor. Based on the monomeric and dimeric DT crystal structures we have determined and the solution studies of others, we present a 5-step structure-based mechanism of intoxication: (1) proteolysis of a disulfide-linked surface loop (residues 186-201) between the catalytic (C) and transmembrane (T) domains; (2) binding of a beta-hairpin loop protruding from the R domain to the DT receptor, leading to receptor-mediated endocytosis; (3) low pH-triggered open monomer formation and exposure of apolar surfaces in the T domain, which insert into the endosomal membrane; (4) translocation of the C domain into the cytosol; and (5) catalysis by the C domain of ADP-ribosylation of elongation factor 2.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California at Los Angeles 90024-1570.