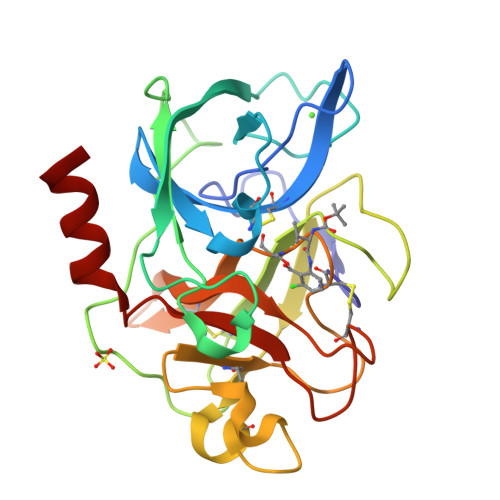
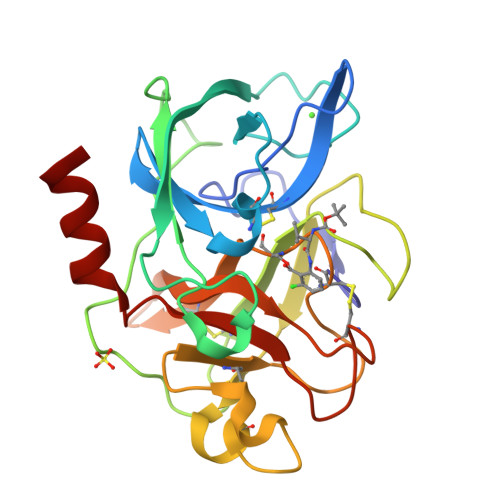
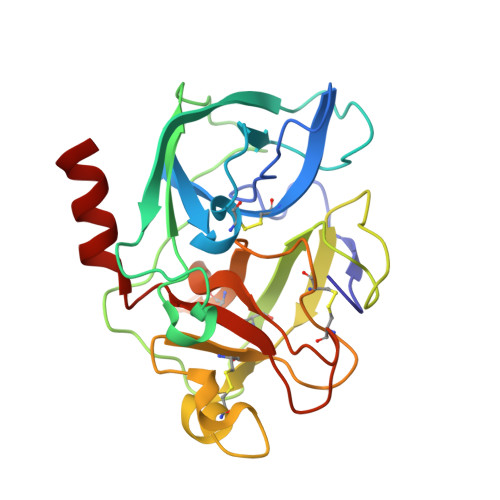
Crystal structures of the complex of porcine pancreatic elastase with two valine-derived benzoxazinone inhibitors.
Radhakrishnan, R., Presta, L.G., Meyer Jr., E.F., Wildonger, R.(1987) J Mol Biology 198: 417-424
- PubMed: 3430613
- DOI: https://doi.org/10.1016/0022-2836(87)90291-9
- Primary Citation of Related Structures:
1INC - PubMed Abstract:
The crystal structures of porcine pancreatic elastase complexed to two similar benzoxazinone inhibitors are reported to 2.09 A and 1.76 A resolution, and refined to conventional R factors of 0.153 and 0.172.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Texas A & M University, College Station 77843.