Full Text |
QUERY: Structure Author = "Shen, H." | MyPDB Login | Search API |
| Search Summary | This query matches 67 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismMore... TaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameMembrane Protein AnnotationSymmetry TypeSCOP Classification | 1 to 25 of 67 Structures Page 1 of 3 Sort by
STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 7-DEAZAGUANINEMiller, D.J., Ravikumar, K., Shen, H., Suh, J.-K., Kerwin, S.M., Robertus, J.D. (2002) J Med Chem 45: 90-98
STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 9-DEAZAGUANINEMiller, D.J., Ravikumar, K., Shen, H., Suh, J.-K., Kerwin, S.M., Robertus, J.D. (2002) J Med Chem 45: 90-98
STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 2,5-DIAMINO-4,6-DIHYDROXYPYRIMIDINE (DDP)Miller, D.J., Ravikumar, K., Shen, H., Suh, J.-K., Kerwin, S.M., Robertus, J.D. (2002) J Med Chem 45: 90-98
STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 8-METHYL-9-OXOGUANINEMiller, D.J., Ravikumar, K., Shen, H., Suh, J.-K., Kerwin, S.M., Robertus, J.D. (2002) J Med Chem 45: 90-98
Yeast F1 ATPase in the absence of bound nucleotidesKabaleeswaran, V., Symersky, J., Shen, H., Walker, J.E., Leslie, A.G.W., Mueller, D.M. (2009) J Biological Chem 284: 10546-10551
Crystal structure of human retinoic X receptor alpha ligand-binding domain complexed with LX0278 and SRC1 peptideZhang, H., Zhang, Y., Shen, H., Chen, J., Li, C., Chen, L., Hu, L., Jiang, H., Shen, X. (2012) PLoS One 7: e31811-e31811
A histone deacetylase from Saccharomyces cerevisiaeZhu, Y., Shen, H., Li, X., Teng, M. (2016) Sci Rep 6: 33905-33905
Crystal Structure Analysis of human CLYBL in complex with free CoASH(2017) Cell 171: 771-782.e11
Crystal Structure Analysis of human CLYBL in complex with propionyl-CoA(2017) Cell 171: 771-782.e11
Crystal Structure Analysis of human CLYBL in apo form(2017) Cell 171: 771-782.e11
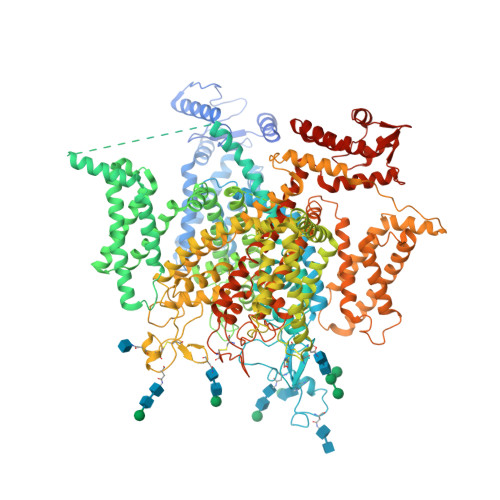
Structure of a eukaryotic voltage-gated sodium channel at near atomic resolutionShen, H., Zhou, Q., Pan, X., Li, Z., Wu, J., Yan, N. (2017) Science 355:
Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6Yu, S.H., Shen, H., Li, X., Mu, W.M. (2018) ACS Catal 8: 10683-10697
Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with DFA-IIIYu, S.H., Shen, H., Li, X., Mu, W.M. (2018) ACS Catal 8: 10683-10697
Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with GF2Yu, S.H., Shen, H., Li, X., Mu, W.M. (2018) ACS Catal 8: 10683-10697
Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 without its lidYu, S.H., Shen, H., Li, X., Mu, W.M. (2018) ACS Catal 8: 10683-10697
Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 wihout its lid in complex with GF2Yu, S.H., Shen, H., Li, X., Mu, W.M. (2018) ACS Catal 8: 10683-10697
Crystal structure of DFA-IIIase mutant C387A from Arthrobacter chlorophenolicus A6Yu, S.H., Shen, H., Li, X., Mu, W.M. (2018) ACS Catal 8: 10683-10697
Crystal structure of mutant C387A of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with DFA-IIIYu, S.H., Shen, H., Li, X., Mu, W.M. (2018) ACS Catal 8: 10683-10697
DHF46 filamentLynch, E.M., Shen, H., Fallas, J.A., Kollman, J.M., Baker, D. (2018) Science 362: 705-709
DHF58 filamentLynch, E.M., Shen, H., Fallas, J.A., Kollman, J.M., Baker, D. (2018) Science 362: 705-709
DHF79 filamentLynch, E.M., Shen, H., Fallas, J.A., Kollman, J.M., Baker, D. (2018) Science 362: 705-709
DHF91 filamentLynch, E.M., Shen, H., Fallas, J.A., Kollman, J.M., Baker, D. (2018) Science 362: 705-709
DHF38 filamentLynch, E.M., Shen, H., Fallas, J.A., Kollman, J.M., Baker, D. (2018) Science 362: 705-709
DHF119 filamentLynch, E.M., Shen, H., Fallas, J.A., Kollman, J.M., Baker, D. (2018) Science 362: 705-709
Structure of human voltage-gated sodium channel Nav1.7 in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (Y1755 up)Shen, H., Liu, D., Lei, J., Yan, N. (2019) Science 363: 1303-1308
1 to 25 of 67 Structures Page 1 of 3 Sort by |