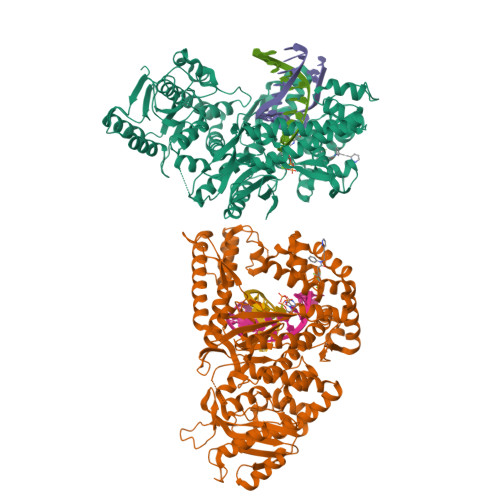
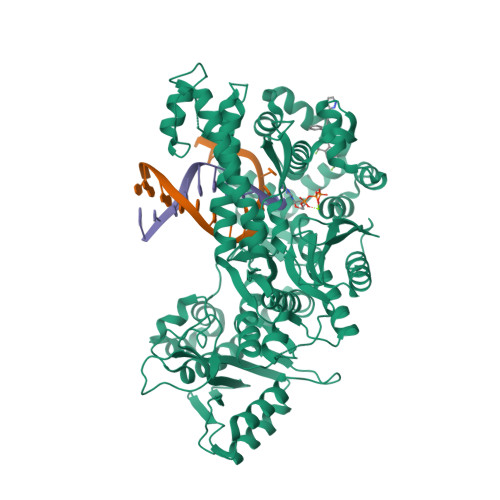
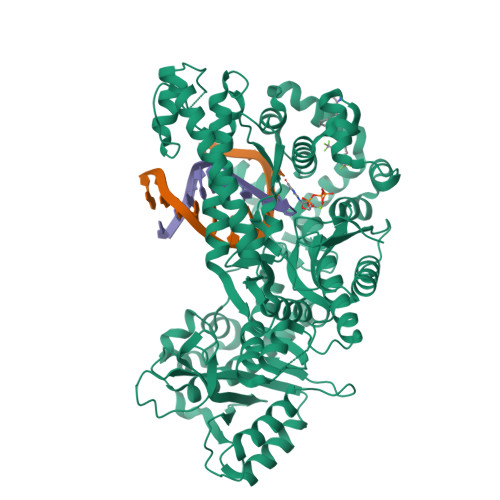
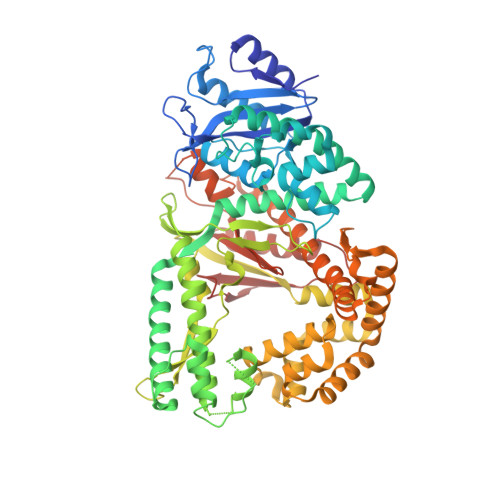
Identification of RP-6685 , an Orally Bioavailable Compound that Inhibits the DNA Polymerase Activity of Pol theta.
Bubenik, M., Mader, P., Mochirian, P., Vallee, F., Clark, J., Truchon, J.F., Perryman, A.L., Pau, V., Kurinov, I., Zahn, K.E., Leclaire, M.E., Papp, R., Mathieu, M.C., Hamel, M., Duffy, N.M., Godbout, C., Casas-Selves, M., Falgueyret, J.P., Baruah, P.S., Nicolas, O., Stocco, R., Poirier, H., Martino, G., Fortin, A.B., Roulston, A., Chefson, A., Dorich, S., St-Onge, M., Patel, P., Pellerin, C., Ciblat, S., Pinter, T., Barabe, F., El Bakkouri, M., Parikh, P., Gervais, C., Sfeir, A., Mamane, Y., Morris, S.J., Black, W.C., Sicheri, F., Gallant, M.(2022) J Med Chem 65: 13198-13215
- PubMed: 36126059
- DOI: https://doi.org/10.1021/acs.jmedchem.2c00998
- Primary Citation of Related Structures:
8E23, 8E24 - PubMed Abstract:
DNA polymerase theta (Polθ) is an attractive synthetic lethal target for drug discovery, predicted to be efficacious against breast and ovarian cancers harboring BRCA-mutant alleles. Here, we describe our hit-to-lead efforts in search of a selective inhibitor of human Polθ (encoded by POLQ). A high-throughput screening campaign of 350,000 compounds identified an 11 micromolar hit, giving rise to the N2-substituted fused pyrazolo series, which was validated by biophysical methods. Structure-based drug design efforts along with optimization of cellular potency and ADME ultimately led to the identification of RP-6685 : a potent, selective, and orally bioavailable Polθ inhibitor that showed in vivo efficacy in an HCT116 BRCA2 -/- mouse tumor xenograft model.
Organizational Affiliation:
Repare Therapeutics, 7171 Frederick-Banting, Building 2, Montréal, Québec H4S 1Z9, Canada.