Polypharmacology of epacadostat: a potent and selective inhibitor of the tumor associated carbonic anhydrases IX and XII.
Angeli, A., Ferraroni, M., Nocentini, A., Selleri, S., Gratteri, P., Supuran, C.T., Carta, F.(2019) Chem Commun (Camb) 55: 5720-5723
- PubMed: 31038135
- DOI: https://doi.org/10.1039/c8cc09568j
- Primary Citation of Related Structures:
6IC2 - PubMed Abstract:
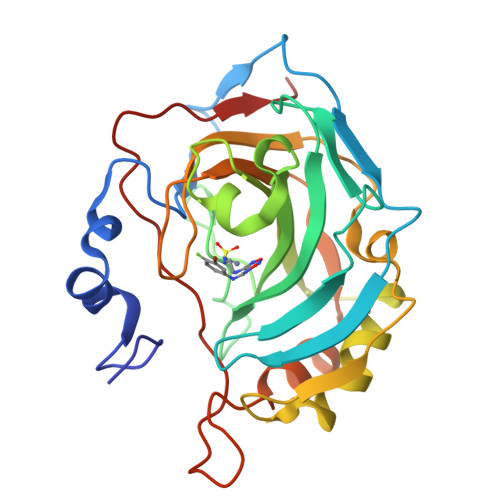
Epacadostat (EPA), a selective indoleamine-2,3-dioxygenase 1 (IDO1) inhibitor, has been investigated in vitro as a human (h) Carbonic Anhydrase Inhibitor (CAI). The kinetic data clearly show, for the first time, EPA to be a highly effective and selective inhibitor for the tumor-associated isoforms hCA IX/XII. We report the high resolution X-ray crystal structure of the EPA-hCA II adduct, and assessed its binding mode to CA IX/XII by means of computational techniques. EPA may exert antitumor effects also due to the potent inhibition of the tumor-associated CAs.
Organizational Affiliation:
University of Florence, NEUROFARBA Dept., Sezione di Farmaceutica e Nutraceutica, Via Ugo Schiff 6, Sesto Fiorentino, 50019, Florence, Italy. claudiu.supuran@unifi.it fabrizio.carta@unifi.it.