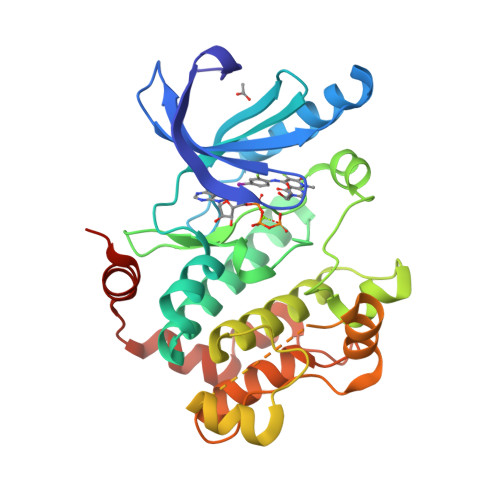
Discovery of TAK-733, a potent and selective MEK allosteric site inhibitor for the treatment of cancer.
Dong, Q., Dougan, D.R., Gong, X., Halkowycz, P., Jin, B., Kanouni, T., O'Connell, S.M., Scorah, N., Shi, L., Wallace, M.B., Zhou, F.(2011) Bioorg Med Chem Lett 21: 1315-1319
- PubMed: 21310613
- DOI: https://doi.org/10.1016/j.bmcl.2011.01.071
- Primary Citation of Related Structures:
3PP1 - PubMed Abstract:
A novel 5-phenylamino-8-methylpyrido[2,3-d]pyrimidine-4,7(3H,8H)-dione series of MEK inhibitors has been developed using structure-based drug design. Lead optimization of this series led to the discovery of TAK-733. This was advanced to Phase I clinical studies for cancer treatment.
Organizational Affiliation:
Takeda San Diego, San Diego, CA 92121, USA.