Selective oxidation of active site aromatic residues in engineered Cu proteins
Uyeda, K.S., Follmer, A.H., Borovik, A.S.(2024) Chem Sci
Experimental Data Snapshot
Starting Model: experimental
View more details
(2024) Chem Sci
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Streptavidin | 159 | Streptomyces avidinii | Mutation(s): 3 | 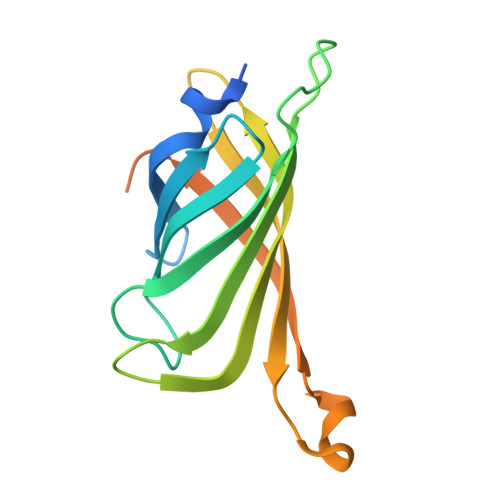 | |
UniProt | |||||
Find proteins for P22629 (Streptomyces avidinii) Explore P22629 Go to UniProtKB: P22629 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P22629 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| QG7 (Subject of Investigation/LOI) Query on QG7 | B [auth A] | N-(2-{bis[(pyridin-2-yl)methyl]amino}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide C24 H32 N6 O2 S GXLJCVSDSFWSLD-FUDKSRODSA-N |  | ||
| CU Query on CU | C [auth A], E [auth A] | COPPER (II) ION Cu JPVYNHNXODAKFH-UHFFFAOYSA-N |  | ||
| ACY (Subject of Investigation/LOI) Query on ACY | D [auth A] | ACETIC ACID C2 H4 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 57.77 | α = 90 |
| b = 57.77 | β = 90 |
| c = 184.25 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MOSFLM | data reduction |
| PHENIX | refinement |
| PHASER | phasing |
| Aimless | data scaling |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM120349 |