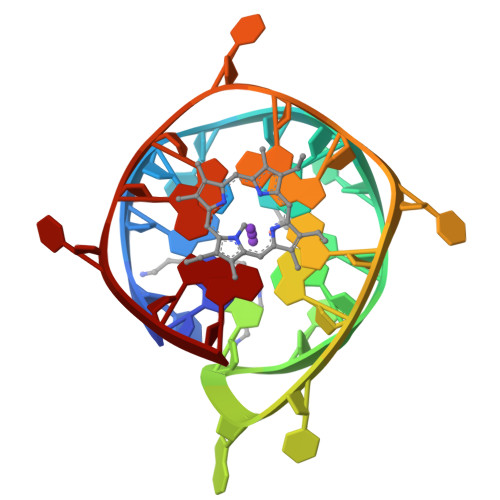
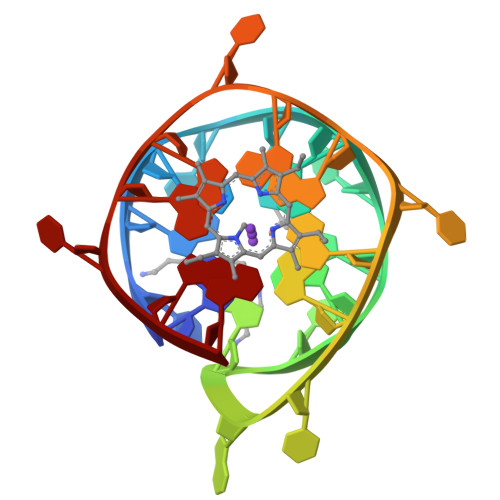
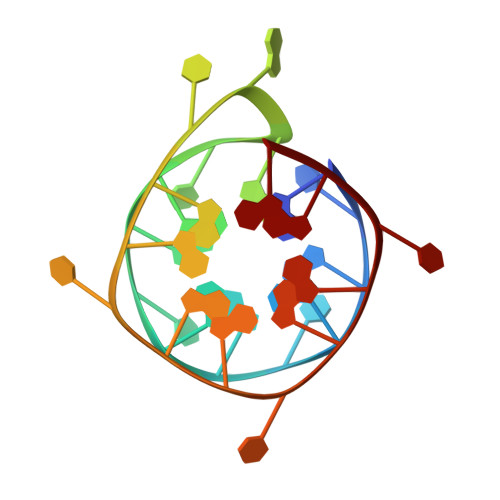
Interaction of N-methylmesoporphyrin IX with a hybrid left-/right-handed G-quadruplex motif from the promoter of the SLC2A1 gene.
Seth, P., Xing, E., Hendrickson, A.D., Li, K., Monsen, R., Chaires, J.B., Neidle, S., Yatsunyk, L.A.(2025) Nucleic Acids Res 53
- PubMed: 39704129
- DOI: https://doi.org/10.1093/nar/gkae1208
- Primary Citation of Related Structures:
8TAA, 9C46 - PubMed Abstract:
Left-handed G-quadruplexes (LHG4s) belong to a class of recently discovered noncanonical DNA structures under the larger umbrella of G-quadruplex DNAs (G4s). The biological relevance of these structures and their ability to be targeted with classical G4 ligands is underexplored. Here, we explore whether the putative LHG4 DNA sequence from the SLC2A1 oncogene promoter maintains its left-handed characteristics upon addition of nucleotides in the 5'- and 3'-direction from its genomic context. We also investigate whether this sequence interacts with a well-established G4 binder, N-methylmesoporphyrin IX (NMM). We employed biophysical and X-ray structural studies to address these questions. Our results indicate that the sequence d[G(TGG)3TGA(TGG)4] (termed here as SLC) adopts a two-subunit, four-tetrad hybrid left-/right-handed G4 (LH/RHG4) topology. Addition of 5'-G or 5'-GG abolishes the left-handed fold in one subunit, while the addition of 3'-C or 3'-CA maintains the original fold. X-ray crystal structure analyses show that SLC maintains the same hybrid LH/RHG4 fold in the solid state and that NMM stacks onto the right-handed subunit of SLC. NMM binds to SLC with a 1:1 stoichiometry and a moderate-to-tight binding constant of 15 μM-1. This work deepens our understanding of LHG4 structures and their binding with traditional G4 ligands.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Swarthmore College, 500 College Ave, Swarthmore, PA, 19081USA.