Insights into the binding of pyrazolopyrimidines to Src kinase
Dello Iacono, L., Kleinboelting, S., Passannanti, R., Fallacara, A.L., Di Pisa, F., Richters, A., Musumeci, F., Rauh, D., Schenone, S., Botta, M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Proto-oncogene tyrosine-protein kinase Src | 286 | Gallus gallus | Mutation(s): 0 Gene Names: SRC EC: 2.7.10.2 | 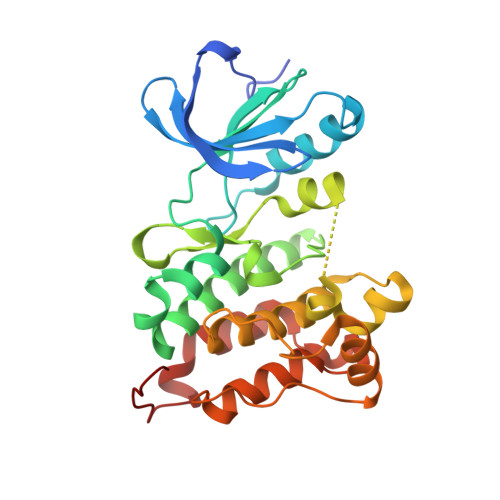 | |
UniProt | |||||
Find proteins for P00523 (Gallus gallus) Explore P00523 Go to UniProtKB: P00523 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00523 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| RCK (Subject of Investigation/LOI) Query on RCK | C [auth A], G [auth B] | ~{N}-[(2-fluorophenyl)methyl]-6-(morpholin-4-ylmethylsulfanyl)-1-(2-phenylethyl)pyrazolo[3,4-d]pyrimidin-4-amine C25 H27 F N6 O S VXDFUYOZJBQCMJ-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | J [auth B], K [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| DMS Query on DMS | F [auth A] | DIMETHYL SULFOXIDE C2 H6 O S IAZDPXIOMUYVGZ-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | D [auth A], E [auth A], H [auth B], I [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 42.26 | α = 101.722 |
| b = 63.42 | β = 90.895 |
| c = 74.93 | γ = 90.008 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Italian Association for Cancer Research | Italy | AIRC IG_4538 |