Discovery of Roblitinib (FGF401) as a Reversible-Covalent Inhibitor of the Kinase Activity of Fibroblast Growth Factor Receptor 4.
Fairhurst, R.A., Knoepfel, T., Buschmann, N., Leblanc, C., Mah, R., Todorov, M., Nimsgern, P., Ripoche, S., Niklaus, M., Warin, N., Luu, V.H., Madoerin, M., Wirth, J., Graus-Porta, D., Weiss, A., Kiffe, M., Wartmann, M., Kinyamu-Akunda, J., Sterker, D., Stamm, C., Adler, F., Buhles, A., Schadt, H., Couttet, P., Blank, J., Galuba, I., Trappe, J., Voshol, J., Ostermann, N., Zou, C., Berghausen, J., Del Rio Espinola, A., Jahnke, W., Furet, P.(2020) J Med Chem 63: 12542-12573
- PubMed: 32930584
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01019
- Primary Citation of Related Structures:
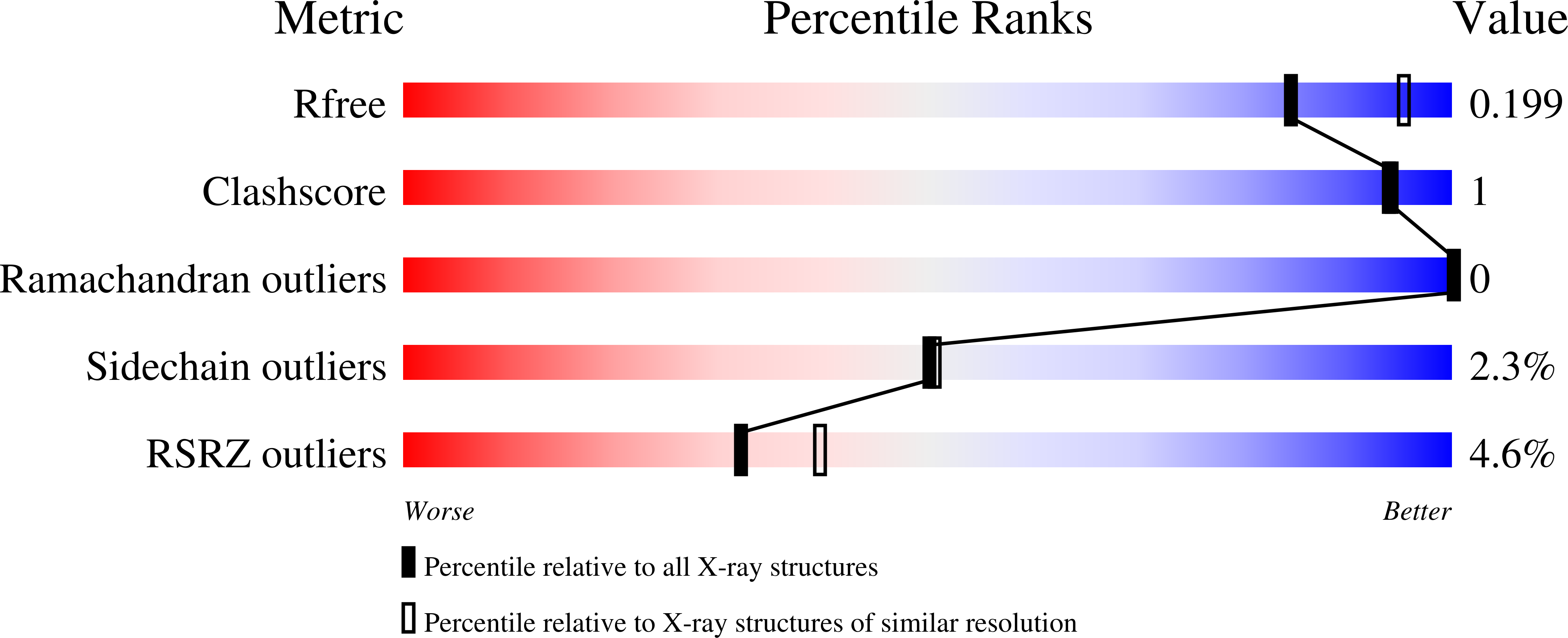
6YI8 - PubMed Abstract:
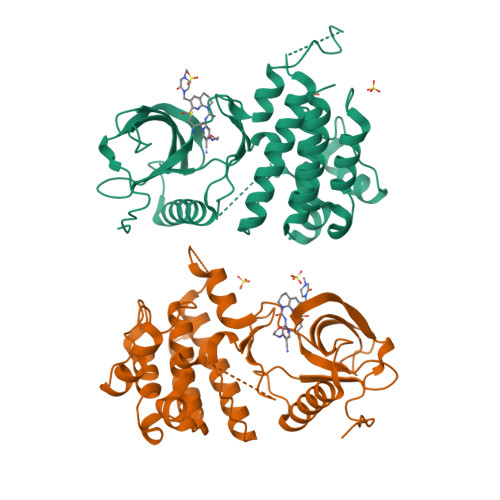
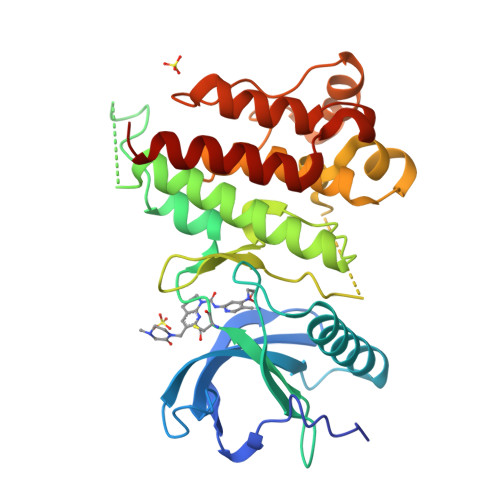
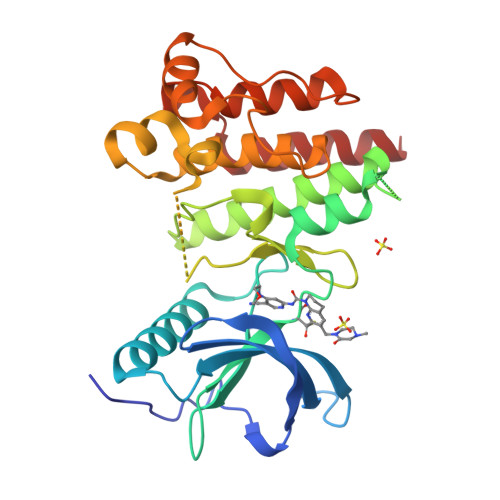
FGF19 signaling through the FGFR4/β-klotho receptor complex has been shown to be a key driver of growth and survival in a subset of hepatocellular carcinomas, making selective FGFR4 inhibition an attractive treatment opportunity. A kinome-wide sequence alignment highlighted a poorly conserved cysteine residue within the FGFR4 ATP-binding site at position 552, two positions beyond the gate-keeper residue. Several strategies for targeting this cysteine to identify FGFR4 selective inhibitor starting points are summarized which made use of both rational and unbiased screening approaches. The optimization of a 2-formylquinoline amide hit series is described in which the aldehyde makes a hemithioacetal reversible-covalent interaction with cysteine 552. Key challenges addressed during the optimization are improving the FGFR4 potency, metabolic stability, and solubility leading ultimately to the highly selective first-in-class clinical candidate roblitinib.
Organizational Affiliation:
Novartis Institutes for BioMedical Research, CH-4002 Basel, Switzerland.