Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Mishra, V., Rathore, I., Arekar, A., Sthanam, L.K., Xiao, H., Kiso, Y., Sen, S., Patankar, S., Gustchina, A., Hidaka, K., Wlodawer, A., Yada, R.Y., Bhaumik, P.(2018) FEBS J 285: 3077-3096
- PubMed: 29943906
- DOI: https://doi.org/10.1111/febs.14598
- Primary Citation of Related Structures:
5YIA, 5YIB, 5YIC, 5YID, 5YIE - PubMed Abstract:
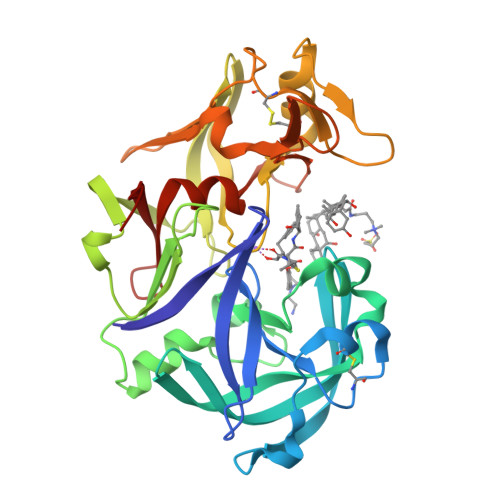
Malaria is a deadly disease killing worldwide hundreds of thousands people each year and the responsible parasite has acquired resistance to the available drug combinations. The four vacuolar plasmepsins (PMs) in Plasmodium falciparum involved in hemoglobin (Hb) catabolism represent promising targets to combat drug resistance. High antimalarial activities can be achieved by developing a single drug that would simultaneously target all the vacuolar PMs. We have demonstrated for the first time the use of soluble recombinant plasmepsin II (PMII) for structure-guided drug discovery with KNI inhibitors. Compounds used in this study (KNI-10742, 10743, 10395, 10333, and 10343) exhibit nanomolar inhibition against PMII and are also effective in blocking the activities of PMI and PMIV with the low nanomolar K i values. The high-resolution crystal structures of PMII-KNI inhibitor complexes reveal interesting features modulating their differential potency. Important individual characteristics of the inhibitors and their importance for potency have been established. The alkylamino analog, KNI-10743, shows intrinsic flexibility at the P2 position that potentiates its interactions with Asp132, Leu133, and Ser134. The phenylacetyl tripeptides, KNI-10333 and KNI-10343, accommodate different ρ-substituents at the P3 phenylacetyl ring that determine the orientation of the ring, thus creating novel hydrogen-bonding contacts. KNI-10743 and KNI-10333 possess significant antimalarial activity, block Hb degradation inside the food vacuole, and show no cytotoxicity on human cells; thus, they can be considered as promising candidates for further optimization. Based on our structural data, novel KNI derivatives with improved antimalarial activity could be designed for potential clinical use. DATABASE: Structural data are available in the PDB under the accession numbers 5YIE, 5YIB, 5YID, 5YIC, and 5YIA.
Organizational Affiliation:
Department of Biosciences and Bioengineering, Indian Institute of Technology Bombay, Powai, Mumbai, India.