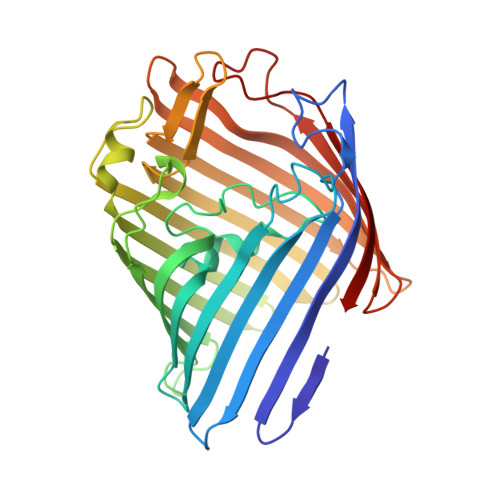
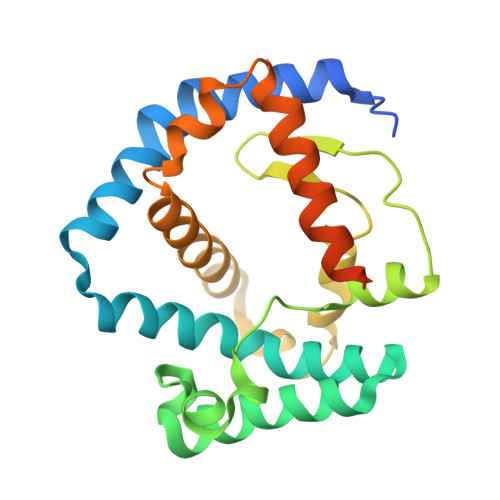
Structural basis for maintenance of bacterial outer membrane lipid asymmetry.
Abellon-Ruiz, J., Kaptan, S.S., Basle, A., Claudi, B., Bumann, D., Kleinekathofer, U., van den Berg, B.(2017) Nat Microbiol 2: 1616-1623
- PubMed: 29038444
- DOI: https://doi.org/10.1038/s41564-017-0046-x
- Primary Citation of Related Structures:
5NUO, 5NUP, 5NUQ, 5NUR - PubMed Abstract:
The Gram-negative bacterial outer membrane (OM) is a unique bilayer that forms an efficient permeation barrier to protect the cell from noxious compounds 1 , 2 . The defining characteristic of the OM is lipid asymmetry, with phospholipids comprising the inner leaflet and lipopolysaccharides comprising the outer leaflet 1-3 . This asymmetry is maintained by the Mla pathway, a six-component system that is widespread in Gram-negative bacteria and is thought to mediate retrograde transport of misplaced phospholipids from the outer leaflet of the OM to the cytoplasmic membrane 4 . The OM lipoprotein MlaA performs the first step in this process via an unknown mechanism that does not require external energy input. Here we show, using X-ray crystallography, molecular dynamics simulations and in vitro and in vivo functional assays, that MlaA is a monomeric α-helical OM protein that functions as a phospholipid translocation channel, forming a ~20-Å-thick doughnut embedded in the inner leaflet of the OM with a central, amphipathic pore. This architecture prevents access of inner leaflet phospholipids to the pore, but allows outer leaflet phospholipids to bind to a pronounced ridge surrounding the channel, followed by diffusion towards the periplasmic space. Enterobacterial MlaA proteins form stable complexes with OmpF/C 5,6 , but the porins do not appear to play an active role in phospholipid transport. MlaA represents a lipid transport protein that selectively removes outer leaflet phospholipids to help maintain the essential barrier function of the bacterial OM.
Organizational Affiliation:
Institute for Cell and Molecular Biosciences, The Medical School, Newcastle University, Newcastle upon Tyne, NE2 4HH, UK.