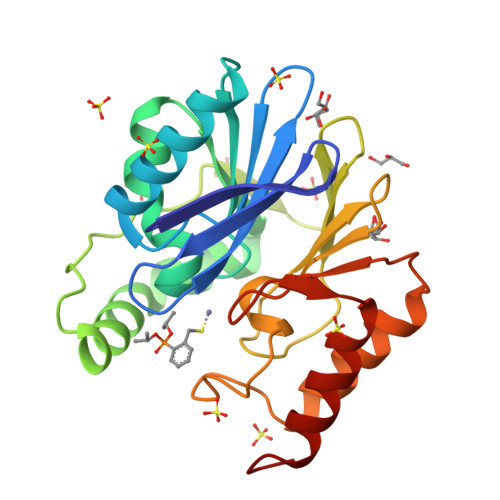
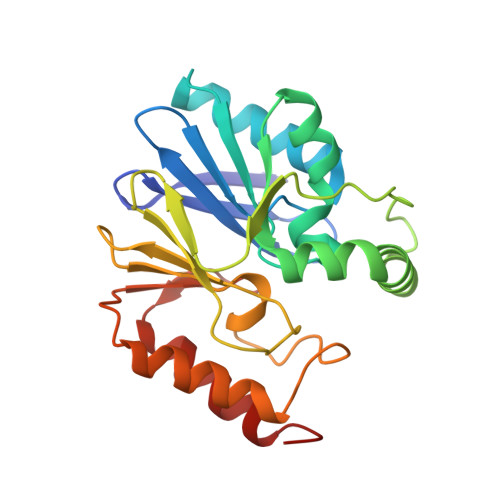
Mercaptophosphonate Compounds as Broad-Spectrum Inhibitors of the Metallo-beta-lactamases
Lassaux, P., Hamel, M., Gulea, M., Delbruck, H., Mercuri, P.S., Horsfall, L., Dehareng, D., Kupper, M., Frere, J.-M., Hoffmann, K., Galleni, M., Bebrone, C.(2010) J Med Chem 53: 4862-4876
- PubMed: 20527888
- DOI: https://doi.org/10.1021/jm100213c
- Primary Citation of Related Structures:
2WRS, 3IOF, 3IOG - PubMed Abstract:
Although commercialized inhibitors of active site serine beta-lactamases are currently used in coadministration with antibiotic therapy, no clinically useful inhibitors of metallo-beta-lactamases (MBLs) have yet been discovered. In this paper, we investigated the inhibitory effect of mercaptophosphonate derivatives against the three subclasses of MBLs (B1, B2, and B3). All 14 tested mercaptophosphonates, with the exception of 1a, behaved as competitive inhibitors for the three subclasses. Apart from 13 and 21, all the mercaptophosphonates tested exhibit a good inhibitory effect on the subclass B2 MBL CphA with low inhibition constants (K(i) < 15 muM). Interestingly, compound 18 turned out to be a potent broad spectrum MBL inhibitor. The crystallographic structures of the CphA-10a and CphA-18 complexes indicated that the sulfur atom of 10a and the phosphonato group of 18 interact with the Zn(2+) ion, respectively. Molecular modeling studies of the interactions between compounds 10a and 18 and the VIM-4 (B1), CphA (B2), and FEZ-1 (B3) enzymes brought to light different binding modes depending on the enzyme and the inhibitor, consistent with the crystallographic structures.
Organizational Affiliation:
Laboratory of Biological Macromolecules, University of Liège, Allée du 6 Août B6, Sart-Tilman, 4000 Liège, Belgium.