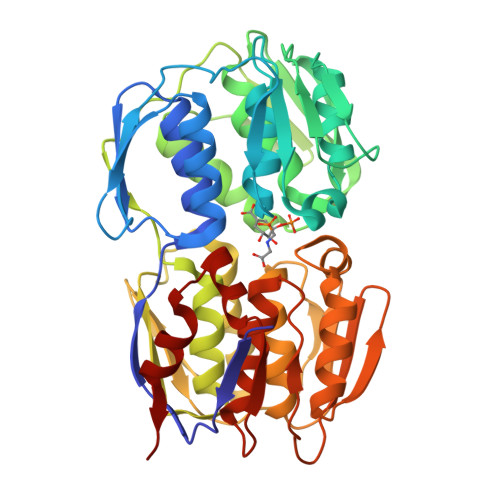
Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
Funke, T., Yang, Y., Han, H., Healy-Fried, M., Olesen, S., Becker, A., Schonbrunn, E.(2009) J Biol Chem 284: 9854-9860
- PubMed: 19211556
- DOI: https://doi.org/10.1074/jbc.M809771200
- Primary Citation of Related Structures:
3FJX, 3FJZ, 3FK0, 3FK1 - PubMed Abstract:
The shikimate pathway enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) is the target of the broad spectrum herbicide glyphosate. The genetic engineering of EPSPS led to the introduction of glyphosate-resistant crops worldwide. The genetically engineered corn lines NK603 and GA21 carry distinct EPSPS enzymes. CP4 EPSPS, expressed in NK603 corn and transgenic soybean, cotton, and canola, belongs to class II EPSPS, glyphosate-insensitive variants of this enzyme isolated from certain Gram-positive bacteria. GA21 corn, on the other hand, was created by point mutations of class I EPSPS, such as the enzymes from Zea mays or Escherichia coli, which are sensitive to low glyphosate concentrations. The structural basis of the glyphosate resistance resulting from these point mutations has remained obscure. We studied the kinetic and structural effects of the T97I/P101S double mutation, the molecular basis for GA21 corn, using EPSPS from E. coli. The T97I/P101S enzyme is essentially insensitive to glyphosate (K(i) = 2.4 mm) but maintains high affinity for the substrate phosphoenolpyruvate (PEP) (K(m) = 0.1 mm). The crystal structure at 1.7-A resolution revealed that the dual mutation causes a shift of residue Gly(96) toward the glyphosate binding site, impairing efficient binding of glyphosate, while the side chain of Ile(97) points away from the substrate binding site, facilitating PEP utilization. The single site T97I mutation renders the enzyme sensitive to glyphosate and causes a substantial decrease in the affinity for PEP. Thus, only the concomitant mutations of Thr(97) and Pro(101) induce the conformational changes necessary to produce catalytically efficient, glyphosate-resistant class I EPSPS.
Organizational Affiliation:
Drug Discovery Department, Moffitt Cancer Center & Research Institute, Tampa, Florida 33612, USA.