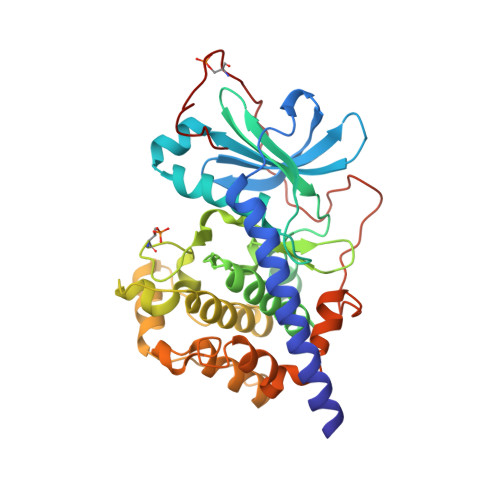
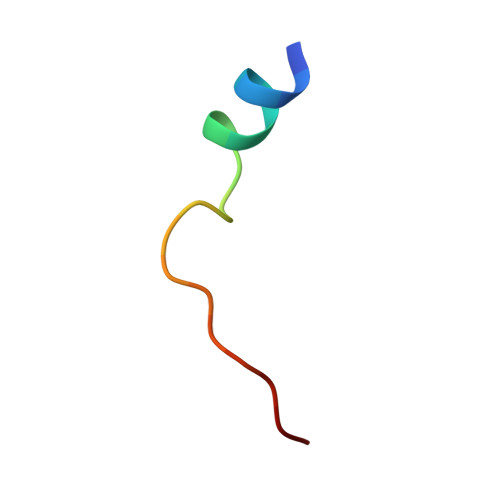
The Typically Disordered N-Terminus of PKA Can Fold as a Helix and Project the Myristoylation Site into Solution
Breitenlechner, C., Engh, R.A., Huber, R., Kinzel, V., Bossemeyer, D., Gassel, M.(2004) Biochemistry 43: 7743-7749
- PubMed: 15196017
- DOI: https://doi.org/10.1021/bi0362525
- Primary Citation of Related Structures:
1SMH - PubMed Abstract:
Protein kinases comprise the major enzyme family critically involved in signal transduction pathways; posttranslational modifications affect their regulation and determine signaling states. The prototype protein kinase A (PKA) possesses an N-terminal alpha-helix (Helix A) that is atypical for kinases and is thus a major distinguishing feature of PKA. Its physiological function may involve myristoylation at the N-terminus and modulation via phosphorylation at serine 10. Here we describe an unusual structure of an unmyristoylated PKA, unphosphorylated at serine 10, with a completely ordered N-terminus. Using standard conditions (e.g., PKI 5-24, ATP site ligand, MEGA-8), a novel 2-fold phosphorylated PKA variant showed the ordered N-terminus in a new crystal packing arrangement. Thus, the critical factor for structuring the N-terminus is apparently the absence of phosphorylation of Ser10. The flexibility of the N-terminus, its myristoylation, and the conformational dependence on the phosphorylation state are consistent with a functional role for myristoylation.
Organizational Affiliation:
Abteilung Strukturforschung, Max-Planck-Institut fuer Biochemie, 82152 Martinsried, Germany. [email protected]